Construction and application of an efficient dual-base editing platform for Bacillus subtilis evolution employing programmable base conversion†
Abstract
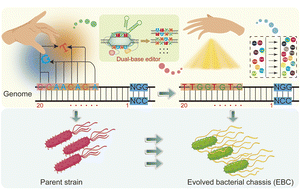
The functionally evolved bacterial chassis is of great importance to manufacture a group of assorted high value-added chemicals, from small molecules to biologically active macromolecules. However, the current evolution frameworks are less efficienct in generating in vivo genomic diversification because of insufficient tunability, rendering limited evolution spacing for chassis. Here, an engineered genomic diversification platform (CRISPR-ABE8e-CDA-nCas9) leveraging a programmable dual-deaminases base editor was fabricated for rapidly evolving bacterial chassis. The dual-base editor was constructed by reprogramming the CRISPR array, nCas9, and cytidine and adenosine deaminase, enabling single or multiple base conversion at the genomic scale by simultaneous C-to-T and A-to-G conversion in vivo. Employing titration of the Cas-deaminase fusion protein, the platform enabled editing any pre-defined genomic loci with tunable conversion efficiency and editable window, generating a repertoire of mutants with highly diversified genomic sequences. Leveraging the genomic diversification platform, we successfully evolved the nisin-resistant capability of Bacillus subtilis through directed evolution of the subunit of lantibiotic ATP-binding cassette. Therefore, our work provides a portable and programmable genomic diversification platform, which is promising to expedite the fabrication of high-performance and robust bacterial chassis used in the development of biomanufacturing and biopharmaceuticals.


 Please wait while we load your content...
Please wait while we load your content...