A photo-oxidation driven proximity labeling strategy enables profiling of mitochondrial proteome dynamics in living cells†
Abstract
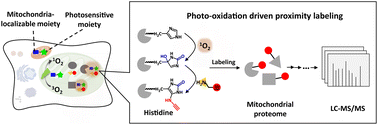
Mapping the proteomic landscape of mitochondria with spatiotemporal precision plays a pivotal role in elucidating the delicate biological functions and complex relationship with other organelles in a variety of dynamic physiological processes which necessitates efficient and controllable chemical tools. We herein report a photo-oxidation driven proximity labeling strategy to profile the mitochondrial proteome by light dependence in living cells with high spatiotemporal resolution. Taking advantage of organelle-localizable organic photoactivated probes generating reactive species and nucleophilic substrates for proximal protein oxidation and trapping, mitochondrial proteins were selectively labeled by spatially limited reactions in their native environment. Integration of photo-oxidation driven proximity labeling and quantitative proteomics facilitated the plotting of the mitochondrial proteome in which up to 310 mitochondrial proteins were identified with a specificity of 64% in HeLa cells. Furthermore, mitochondrial proteome dynamics was deciphered in drug resistant Huh7 and LPS stimulated HMC3 cells which were hard-to-transfect. A number of differential proteins were quantified which were intimately linked to critical processes and provided insights into the related molecular mechanisms of drug resistance and neuroinflammation in the perspective of mitochondria. The photo-oxidation driven proximity labeling strategy offers solid technical support to a highly precise proteomic platform in time and finer space for more knowledge of subcellular biology.


 Please wait while we load your content...
Please wait while we load your content...