How well does molecular simulation reproduce environment-specific conformations of the intrinsically disordered peptides PLP, TP2 and ONEG?†
Abstract
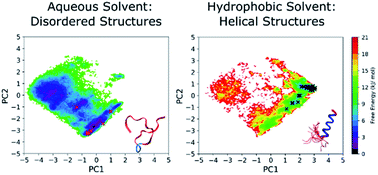
Understanding the conformational ensembles of intrinsically disordered proteins and peptides (IDPs) in their various biological environments is essential for understanding their mechanisms and functional roles in the proteome, leading to a greater knowledge of, and potential treatments for, a broad range of diseases. To determine whether molecular simulation is able to generate accurate conformational ensembles of IDPs, we explore the structural landscape of the PLP peptide (an intrinsically disordered region of the proteolipid membrane protein) in aqueous and membrane-mimicking solvents, using replica exchange with solute scaling (REST2), and examine the ability of four force fields (ff14SB, ff14IDPSFF, CHARMM36 and CHARMM36m) to reproduce literature circular dichroism (CD) data. Results from variable temperature (VT) 1H and Rotating frame Overhauser Effect SpectroscopY (ROESY) nuclear magnetic resonance (NMR) experiments are also presented and are consistent with the structural observations obtained from the simulations and CD. We also apply the optimum simulation protocol to TP2 and ONEG (a cell-penetrating peptide (CPP) and a negative control peptide, respectively) to gain insight into the structural differences that may account for the observed difference in their membrane-penetrating abilities. Of the tested force fields, we find that CHARMM36 and CHARMM36m are best suited to the study of IDPs, and accurately predict a disordered to helical conformational transition of the PLP peptide accompanying the change from aqueous to membrane-mimicking solvents. We also identify an α-helical structure of TP2 in the membrane-mimicking solvents and provide a discussion of the mechanistic implications of this observation with reference to the previous literature on the peptide. From these results, we recommend the use of CHARMM36m with the REST2 protocol for the study of environment-specific IDP conformations. We believe that the simulation protocol will allow the study of a broad range of IDPs that undergo conformational transitions in different biological environments.


 Please wait while we load your content...
Please wait while we load your content...