Learning the structure–activity relationship (SAR) of the Wittig reaction from genetically-encoded substrates†
Abstract
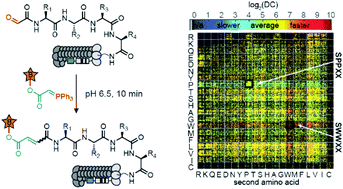
The Wittig reaction can be used for late stage functionalization of proteins and peptides to ligate glycans, pharmacophores, and many other functionalities. In this manuscript, we modified 160 000 N-terminal glyoxaldehyde peptides displayed on phage with the Wittig reaction by using a biotin labeled ylide under conditions that functionalize only 1% of the library population. Deep-sequencing of the biotinylated and input populations estimated the rate of conversion for each sequence. This “deep conversion” (DC) from deep sequencing correlates with rate constants measured by HPLC. Peptide sequences with fast and slow reactivity highlighted the critical role of primary backbone amides (N–H) in accelerating the rate of the aqueous Wittig reaction. Experimental measurement of reaction rates and density functional theory (DFT) computation of the transition state geometries corroborated this relationship. We also collected deep-sequencing data to build structure–activity relationship (SAR) models that can predict the DC value of the Wittig reaction. By using these data, we trained two classifier models based on gradient boosted trees. These classifiers achieved area under the ROC (receiver operating characteristic) curve (ROC AUC) of 81.2 ± 0.4 and 73.7 ± 0.8 (90–92% accuracy) in determining whether a sequence belonged to the top 5% or the bottom 5% in terms of its reactivity. This model can suggest new peptides never observed experimentally with ‘HIGH’ or ‘LOW’ reactivity. Experimental measurement of reaction rates for 11 new sequences corroborated the predictions for 8 of them. We anticipate that phage-displayed peptides and related mRNA or DNA-displayed substrates can be employed in a similar fashion to study the substrate scope and mechanisms of many other chemical reactions.


 Please wait while we load your content...
Please wait while we load your content...