Arrow pushing in RNA modification sequencing
Abstract
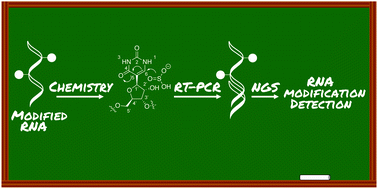
Methods to accurately determine the location and abundance of RNA modifications are critical to understanding their functional role. In this review, we describe recent efforts in which chemical reactivity and next-generation sequencing have been integrated to detect modified nucleotides in RNA. For eleven exemplary modifications, we detail chemical, enzymatic, and metabolic labeling protocols that can be used to differentiate them from canonical nucleobases. By emphasizing the molecular rationale underlying these detection methods, our survey highlights new opportunities for chemistry to define the role of RNA modifications in disease.
- This article is part of the themed collection: Nucleic Acids


 Please wait while we load your content...
Please wait while we load your content...