RPnet: a reverse-projection-based neural network for coarse-graining metastable conformational states for protein dynamics†
Abstract
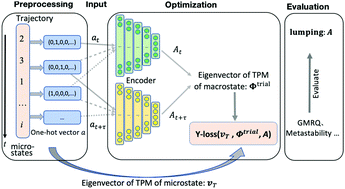
The Markov State Model (MSM) is a powerful tool for modeling long timescale dynamics based on numerous short molecular dynamics (MD) simulation trajectories, which makes it a useful tool for elucidating the conformational changes of biological macromolecules. By partitioning the phase space into discretized states and estimating the probabilities of inter-state transitions based on short MD trajectories, one can construct a kinetic network model that could be used to extrapolate long-timescale kinetics if the Markovian condition is met. However, meeting the Markovian condition often requires hundreds or even thousands of states (microstates), which greatly hinders the comprehension of the conformational dynamics of complex biomolecules. Kinetic lumping algorithms can coarse grain numerous microstates into a handful of metastable states (macrostates), which would greatly facilitate the elucidation of biological mechanisms. In this work, we have developed a reverse-projection-based neural network (RPnet) to lump microstates into macrostates, by making use of a physics-based loss function that is based on the projection operator framework of conformational dynamics. By recognizing that microstate and macrostate transition modes can be related through a projection process, we have developed a reverse-projection scheme to directly compare the microstate and macrostate dynamics. Based on this reverse-projection scheme, we designed a loss function that allows the effective assessment of the quality of a given kinetic lumping. We then make use of a neural network to efficiently minimize this loss function to obtain an optimized set of macrostates. We have demonstrated the power of our RPnet in analyzing the dynamics of a numerical 2D potential, alanine dipeptide, and the clamp opening of an RNA polymerase. In all these systems, we have illustrated that our method could yield comparable or better results than competing methods in terms of state partitioning and reproduction of slow dynamics. We expect that our RPnet holds promise in analyzing the conformational dynamics of biological macromolecules.
- This article is part of the themed collection: 2022 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...