Generation of 3D cellular spheroids using DNA modified cell receptors and programmable DNA interactions†
Abstract
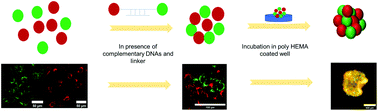
3D culture is known to provide more faithful tissue models than 2D culture, and thus it is a valuable tool for in vitro evaluation of biological models. However, many cell lines are unable to form desired 3D spheroids by traditional methods because the naturally occurring cell–cell adhesion is too weak. Here, we present a method to produce 3D cell spheroids by using DNA-mediated assembly. We first demonstrate an Affinity Mediated Photoconjugation Approach (AMCP) to covalently modify cell receptors with affibody–streptavidin fusion proteins, where the affibody chemically crosslinks to cell expressed EGFR and the streptavidin is used to attach DNA strands. The DNA conjugated cells were then mixed with complementary DNA ‘linker strands’ to impart cell–cell interactions. When incubated in wells coated with non-adhesive polymers, cells formed dense spherical aggregates larger than 500 microns in diameter. Each of these studies was carried out using human breast cancer cells (MBA-MB-468), aneuploid human keratinocytes (HaCaT), and human colon cancer cells (Caco-2). Without either DNA on the cells or in solution as linkers, no cell spheroids were observed. After 96 h of incubation, the cultured DNA assembled spheroids were found to be mechanically stable enough to be handled easily for further analysis and confocal imaging. The findings suggest that the proposed DNA assembly method can be considered as an attractive strategy for assembling cells into stable spheroids.


 Please wait while we load your content...
Please wait while we load your content...