Developments in toxicogenomics: understanding and predicting compound-induced toxicity from gene expression data
Abstract
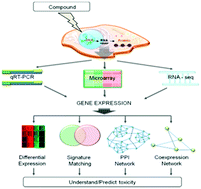
The toxicogenomics field aims to understand and predict toxicity by using ‘omics’ data in order to study systems-level responses to compound treatments. In recent years there has been a rapid increase in publicly available toxicological and ‘omics’ data, particularly gene expression data, and a corresponding development of methods for its analysis. In this review, we summarize recent progress relating to the analysis of RNA-Seq and microarray data, review relevant databases, and highlight recent applications of toxicogenomics data for understanding and predicting compound toxicity. These include the analysis of differentially expressed genes and their enrichment, signature matching, methods based on interaction networks, and the analysis of co-expression networks. In the future, these state-of-the-art methods will likely be combined with new technologies, such as whole human body models, to produce a comprehensive systems-level understanding of toxicity that reduces the necessity of in vivo toxicity assessment in animal models.


 Please wait while we load your content...
Please wait while we load your content...