Mechanistic insights into peptide and ligand binding of the ATAD2-bromodomain via atomistic simulations disclosing a role of induced fit and conformational selection†
Abstract
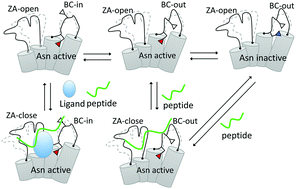
ATAD2 has emerged as a promising bromodomain (BRD)-containing therapeutic drug target in multiple human cancers. However, recent druggability assessment studies predicted ATAD2's BRD as a target ‘difficult to drug’ because its binding pocket possesses structural features that are unfeasible for ligand binding. Here, by using all-atom molecular dynamics simulations and an advanced metadynamics method, we demonstrate a dynamic view of the binding pocket features which can hardly be obtained from the “static” crystal data. The most important features disclosed from our simulation data, include: (1) a distinct ‘open-to-closed’ conformational switch of the ZA loop region in the context of peptide or ligand binding, akin to the induced fit mechanism of molecular recognition, (2) a dynamic equilibrium of the BC loop “in” and “out” conformations, highlighting a role in the conformational selection mechanism for ligand binding, and (3) a new binding region identified distal to the histone-binding pocket that might have implications in bromodomain biology and in inhibitor development. Moreover, based on our simulation results, we propose a model for an “auto-regulatory” mechanism of ATAD2's BRD for histone binding. Overall, the results of this study will not only have implications in bromodomain biology but also provide a theoretical basis for the discovery of new ATAD2's BRD inhibitors.
- This article is part of the themed collection: 2018 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...