Phosphoramidate hydrolysis catalyzed by human histidine triad nucleotide binding protein 1 (hHint1): a cluster-model DFT computational study†
Abstract
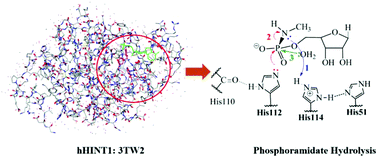
As a member of the histidine triad (HIT) protein superfamily, human histidine triad nucleotide binding protein 1 (hHint1) serves as an efficient enzyme in the hydrolysis of phosphoramidate. In particular, hHint1 has been utilized to activate nucleotide prodrugs (proTides). Understanding the mechanism of hHint1 will aid in the future design of proTides. Density functional theory (DFT) computations on a 228-atom cluster active-site model were performed to investigate the hydrolysis mechanism of a phosphoramidate substrate. The overall proposed mechanism included the key involvement of the histidine triad as a proton shuttle. Protonated methylphosphoramidate was first formed by proton transfer of protonated His114 species. A penta-coordinated phosphoryl intermediate, protonated methylphosphorodiamidate, was generated by a nucleophilic attack of His112. After the release of amine and the generation of a phosphorylated histidine intermediate, the nucleophilic attack of an active-site water produced a hydrolyzed intermediate that subsequently transferred a proton back to His114. A rate-determining fully associative pathway with a free energy of activation of 21.7 kcal mol−1 formed the penta-coordinated phosphoryl intermediate. A non-rate determining associative-interchange transition state was involved in the formation of transient tetra-coordinated phosphoryl intermediate. The overall hydrolysis was favorable by −16.1 kcal mol−1.


 Please wait while we load your content...
Please wait while we load your content...