Molecular dynamics simulations reveal disruptive self-assembly in dynamic peptide libraries†
Abstract
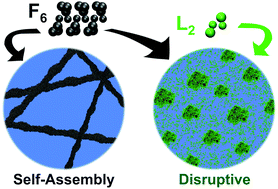
There is significant interest in the use of unmodified self-assembling peptides as building blocks for functional, supramolecular biomaterials. Recently, dynamic peptide libraries (DPLs) have been proposed to select self-assembling materials from dynamically exchanging mixtures of dipeptide inputs in the presence of a nonspecific protease enzyme, where peptide sequences are selected and amplified based on their self-assembling tendencies. It was shown that the results of the DPL of mixed sequences (e.g. starting from a mixture of dileucine, L2, and diphenylalanine, F2) did not give the same outcome as the separate L2 and F2 libraries (which give rise to the formation of F6 and L6), implying that interactions between these sequences could disrupt the self-assembly. In this study, coarse grained molecular dynamics (CG-MD) simulations are used to understand the DPL results for F2, L2 and mixed libraries. CG-MD simulations demonstrate that interactions between precursors can cause the low formation yield of hexapeptides in the mixtures of dipeptides and show that this ability to disrupt is influenced by the concentration of the different species in the DPL. The disrupting self-assembly effect between the species in the DPL is an important effect to take into account in dynamic combinatorial chemistry as it affects the possible discovery of new materials. This work shows that combined computational and experimental screening can be used complementarily and in combination providing a powerful means to discover new supramolecular peptide nanostructures.
- This article is part of the themed collection: Peptide Materials


 Please wait while we load your content...
Please wait while we load your content...