Simultaneous NMR characterisation of multiple minima in the free energy landscape of an RNA UUCG tetraloop†
Abstract
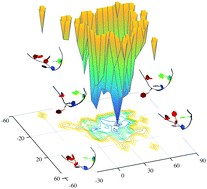
RNA molecules in solution tend to undergo structural fluctuations of relatively large amplitude and to populate a range of different conformations some of which with low populations. It is still very challenging, however, to characterise the structures of these low populated states and to understand their functional roles. In the present study, we address this problem by using NMR residual dipolar couplings (RDCs) as structural restraints in replica-averaged metadynamics (RAM) simulations. By applying this approach to a 14-mer RNA hairpin containing the prototypical UUCG tetraloop motif, we show that it is possible to construct the free energy landscape of this RNA molecule. This free energy landscapes reveals the surprisingly rich dynamics of the UUCG tetraloop and identifies the multiple substates that exist in equilibrium owing to thermal fluctuations. The approach that we present is general and can be applied to the study of the free energy landscapes of other RNA or RNA-protein systems.


 Please wait while we load your content...
Please wait while we load your content...