Signatures of protein thermal denaturation and local hydrophobicity in domain specific hydration behavior: a comparative molecular dynamics study†
Abstract
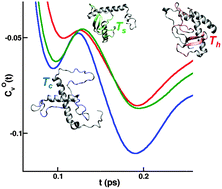
We investigate, using atomistic molecular dynamics simulations, the association of surface hydration accompanying local unfolding in the mesophilic protein Yfh1 under a series of thermal conditions spanning its cold and heat denaturation temperatures. The results are benchmarked against the thermally stable protein, Ubq, and behavior at the maximum stability temperature. Local unfolding in Yfh1, predominantly in the beta sheet regions, is in qualitative agreement with recent solution NMR studies; the corresponding Ubq unfolding is not observed. Interestingly, all domains, except for the beta sheet domains of Yfh1, show increased effective surface hydrophobicity with increase in temperature, as reflected by the density fluctuations of the hydration layer. Velocity autocorrelation functions (VACF) of oxygen atoms of water within the hydration layers and the corresponding vibrational density of states (VDOS) are used to characterize alteration in dynamical behavior accompanying the temperature dependent local unfolding. Enhanced caging effects accompanying transverse oscillations of the water molecules are found to occur with the increase in temperature preferentially for the beta sheet domains of Yfh1. Helical domains of both proteins exhibit similar trends in VDOS with changes in temperature. This work demonstrates the existence of key signatures of the local onset of protein thermal denaturation in solvent dynamical behavior.
- This article is part of the themed collection: Chemical Biology in Molecular BioSystems

 Please wait while we load your content...
Please wait while we load your content...