The relationships among host transcriptional responses reveal distinct signatures underlying viral infection-disease associations†
Abstract
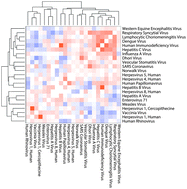
Genome-scale DNA microarrays and computational biology facilitate new understanding of viral infections at the system level. Recent years have witnessed a major shift from microorganism-centric toward host-oriented characterization and categorization of viral infections and infection related diseases. We established host transcriptional response (HTR) relationships among 23 different types of human viral pathogens based on calculating HTR similarities using computational integration of 587 public available gene expression profiles. We further identified five virus clusters that show consensus internal HTRs and defined cluster signatures using common dysregulated genes. Individual cluster signature genes were distinguished from one another, and functional analysis revealed common and specific host cellular bioprocesses and signaling pathways involved in confronting viral infections. Through literature investigation and support from epidemiological studies, these were confirmed to be important gene factors associating viral infections with cluster-common and cluster-specific non-infectious human disease(s). Our analyses were the first to feature differential HTRs to viral infections as clusters, and they present a new perspective for understanding infection-disease associations and the underlying pathogeneses.

 Please wait while we load your content...
Please wait while we load your content...