Molecular basis for competitive solvation of the Burkholderia cepacia lipase by sorbitol and urea
Abstract
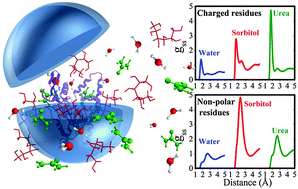
Increasing the stability of proteins is important for their application in industrial processes. In the intracellular environment many small molecules, called osmolytes, contribute to protein stabilization under physical or chemical stress. Understanding the nature of the interactions of these osmolytes with proteins can help the design of solvents and mutations to increase protein stability in extracellular media. One of the most common stabilizing osmolyes is sorbitol and one of the most common chemical denaturants is urea. In this work, we use molecular dynamics simulations to obtain a detailed picture of the solvation of the Burkholderia cepacia lipase (BCL) in the presence of the protecting osmolyte sorbitol and of the urea denaturant. We show that both sorbitol and urea compete with water for interactions with the protein surface. Overall, sorbitol promotes the organization of water in the first solvation shell and displaces water from the second solvation shell, while urea causes opposite effects. These effects are, however, highly heterogeneous among residue types. For instance, the depletion of water from the first protein solvation shell by urea can be traced down essentially to the side chain of negatively charged residues. The organization of water in the first solvation shell promoted by sorbitol occurs at polar (but not charged) residues, where the urea effect is minor. By contrast, sorbitol depletes water from the second solvation shell of polar residues, while urea promotes water organization at the same distances. The interactions of urea with negatively charged residues are insensitive to the presence of sorbitol. This osmolyte removes water and urea particularly from the second solvation shell of polar and non-polar residues. In summary, we provide a comprehensive description of the diversity of protein–solvent interactions, which can guide further investigations on the stability of proteins in non-conventional media, and assist solvent and protein design.

 Please wait while we load your content...
Please wait while we load your content...