Inter-helical conformational preferences of HIV-1 TAR-RNA from maximum occurrence analysis of NMR data and molecular dynamics simulations†
Abstract
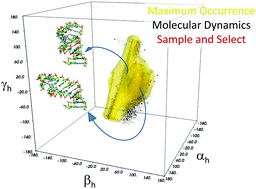
Detecting conformational heterogeneity in biological macromolecules is a key for the understanding of their biological function. We here provide a comparison between two independent approaches to assess conformational heterogeneity: molecular dynamics simulations, performed without inclusion of any experimental data, and maximum occurrence (MaxOcc) distribution over the topologically available conformational space. The latter only reflects the extent of the averaging and identifies regions which are most compliant with the experimentally measured NMR Residual Dipolar Couplings (RDCs). The analysis was performed for the HIV-1 TAR RNA, consisting of two helical domains connected by a flexible bulge junction, for which four sets of RDCs were available as well as an 8.2 μs all-atom molecular dynamics simulation. A sample and select approach was previously applied to extract from the molecular dynamics trajectory conformational ensembles in agreement with the four sets of RDCs. The MaxOcc analysis performed here identifies the most likely sampled region in the conformational space of the system which, strikingly, overlaps well with the structures independently sampled in the molecular dynamics calculations and even better with the RDC selected ensemble.
- This article is part of the themed collection: Exploring the conformational heterogeneity of biomolecules: theory and experiments

 Please wait while we load your content...
Please wait while we load your content...