Mass spectrometry of modified RNAs: recent developments
Abstract
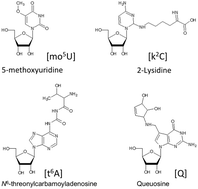
A common feature of ribonucleic acids (RNAs) is that they can undergo a variety of chemical modifications. As nearly all of these chemical modifications result in an increase in the mass of the canonical nucleoside, mass spectrometry has long been a powerful approach for identifying and characterizing modified RNAs. Over the past several years, significant advances have been made in method development and software for interpreting tandem mass spectra resulting in approaches that can yield qualitative and quantitative information on RNA modifications, often at the level of sequence specificity. We discuss these advances along with instrumentation developments that have increased our ability to extract such information from relatively complex biological samples. With the increasing interest in how these modifications impact the epitranscriptome, mass spectrometry will continue to play an important role in bioanalytical investigations revolving around RNA.
 Please wait while we load your content...
Please wait while we load your content...