Experimental design, modeling and optimization of polyplex formation between DNA oligonucleotides and branched polyethylenimine†
Abstract
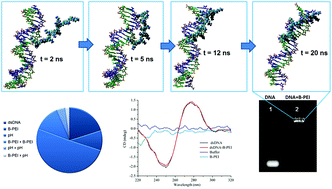
The complexes formed by DNA and polycations have received great attention owing to their potential application in gene therapy. In this study, the binding efficiency between double-stranded oligonucleotides (dsDNA) and branched polyethylenimine (B-PEI) has been quantified by processing of the images captured from the gel electrophoresis assays. The central composite experimental design has been employed to investigate the effects of controllable factors on the binding efficiency. On the basis of experimental data and the response surface methodology, a multivariate regression model has been constructed and statistically validated. The model has enabled us to predict the binding efficiency depending on experimental factors, such as concentrations of dsDNA and B-PEI as well as the initial pH of solution. The optimization of the binding process has been performed using simplex and gradient methods. The optimal conditions determined for polyplex formation have yielded a maximal binding efficiency close to 100%. In order to reveal the mechanism of complex formation at the atomic-scale, a molecular dynamic simulation has been carried out. According to the computation results, B-PEI amine hydrogen atoms have interacted with oxygen atoms from dsDNA phosphate groups. These interactions have led to the formation of hydrogen bonds between macromolecules, stabilizing the polyplex structure.
- This article is part of the themed collection: Multivalent Biomolecular Recognition

 Please wait while we load your content...
Please wait while we load your content...