Structural and functional insights into CARDs of zebrafish (Danio rerio) NOD1 and NOD2, and their interaction with adaptor protein RIP2†
Abstract
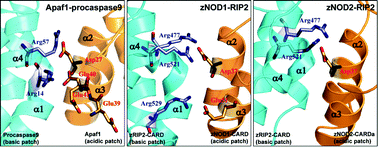
Nucleotide-binding and oligomerization domain-containing protein 1 (NOD1) and NOD2 are cytosolic pattern-recognition receptors (PRRs) composed of an N-terminal caspase activation and recruitment domain (CARD), a central NACHT domain and C-terminal leucine-rich repeats (LRRs). They play a vital role in innate immune signaling by activating the NF-κB pathway via recognition of peptidoglycans by LRRs, and ATP-dependent self-oligomerization of NACHT followed by downstream signaling. After oligomerization, CARD/s play a crucial role in activating downstream signaling via the adaptor molecule, RIP2. Due to the inadequacy of experimental 3D structures of CARD/s of NOD2 and RIP2, and results from differential experimental setups, the RIP2-mediated CARD–CARD interaction has remained as a contradictory statement. We employed a combinatorial approach involving protein modeling, docking, molecular dynamics simulation, and binding free energy calculation to illuminate the molecular mechanism that shows the possible involvement of either the acidic or basic patch of zebrafish NOD1/2–CARD/a and RIP2–CARD in CARD–CARD interaction. Herein, we have hypothesized ‘type-I’ mode of CARD–CARD interaction in NOD1 and NOD2, where NOD1/2–CARD/a involve their acidic surfaces to interact with RIP2. Asp37 and Glu51 (of NOD1) and Arg477, Arg521 and Arg529 (of RIP2) were identified to be crucial for NOD1–RIP2 interaction. However, in NOD2–RIP2, Asp32 (of NOD2) and Arg477 and Arg521 (of RIP2) were anticipated to be significant for downstream signaling. Furthermore, we found that strong electrostatic contacts and salt bridges are crucial for protein–protein interactions. Altogether, our study has provided novel insights into the RIP2-mediated CARD–CARD interaction in zebrafish NOD1 and NOD2, which will be helpful to understand the molecular basis of the NOD1/2 signaling mechanism.

 Please wait while we load your content...
Please wait while we load your content...