Understanding the impact of Fc glycosylation on its conformational changes by molecular dynamics simulations and bioinformatics†
Abstract
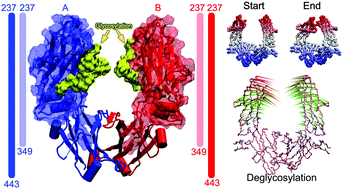
N-linked glycosylation of Fc at N297 plays an important role in its effector function, aberrance of which would cause disease pathogenesis. Here, we performed all-atom molecular dynamics simulations to explore the effects of Fc glycosylation on its dynamics behaviors. Firstly, equilibrium simulations suggested that Fc deglycosylation was able to induce residual flexibility in its CH2 domain. Besides, the free energy landscape revealed three minimum energy wells in deglycosylated Fc, representing its “open”, “semi-closed” and “closed” states. However, we could only observe the “open” state of glycosylated Fc. Supportively, principal component analysis emphasized the prominent motion of delyclosylated Fc and dynamically depicted how it changed from the “open” state to its “closed” state. Secondly, we studied the recognition mechanism of the Fc binding to its partners. Energy decomposition analysis identified key residues of Fc to recognize its two partners P13 and P34. Evidently, electrostatic potential surfaces showed that electrostatic attraction helped to stabilize the interaction between Fc and its partners. Also, relative binding free energies explained different binding affinities in Fc–P13 and Fc–P34. Collectively, these results together provided the structural basis for understanding conformational changes of deglycosylated Fc and the recognition mechanism of the Fc binding to its partners.

 Please wait while we load your content...
Please wait while we load your content...