Naïve Bayes classifier predicts functional microRNA target interactions in colorectal cancer†
Abstract
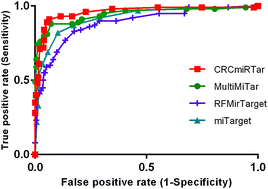
Alterations in the expression of miRNAs have been extensively characterized in several cancers, including human colorectal cancer (CRC). Recent publications provide evidence for tissue-specific miRNA target recognition. Several computational methods have been developed to predict miRNA targets; however, all of these methods assume a general pattern underlying these interactions and therefore tolerate reduced prediction accuracy and a significant number of false predictions. The motivation underlying the presented work was to unravel the relationship between miRNAs and their target mRNAs in CRC. We developed a novel computational algorithm for miRNA–target prediction in CRC using a Naïve Bayes classifier. The algorithm, which is referred to as CRCmiRTar, was trained with data from validated miRNA target interactions in CRC and other cancer entities. Furthermore, we identified a set of position-based, sequence, structural, and thermodynamic features that identify CRC-specific miRNA target interactions. Evaluation of the algorithm showed a significant improvement of performance with respect to AUC, and sensitivity, compared to other widely used algorithms based on machine learning. Based on miRNA and gene expression profiles in CRC tissues with similar clinical and pathological features, our classifier predicted 204 functional interactions, which involve 11 miRNAs and 41 mRNAs in this cancer entity. While the approach is here validated for CRC, the implementation of disease-specific miRNA target prediction algorithms can be easily adopted for other applications too. The identification of disease-specific miRNA target interactions may also facilitate the identification of potential drug targets.

 Please wait while we load your content...
Please wait while we load your content...