Dynamic metabolic models in context: biomass backtracking†
Abstract
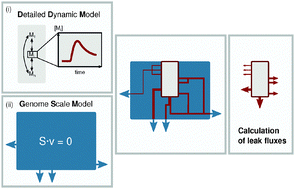
Mathematical modeling has proven to be a powerful tool to understand and predict functional and regulatory properties of metabolic processes. High accuracy dynamic modeling of individual pathways is thereby opposed by simplified but genome scale constraint based approaches. A method that links these two powerful techniques would greatly enhance predictive power but is so far lacking. We present biomass backtracking, a workflow that integrates the cellular context in existing dynamic metabolic models via stoichiometrically exact drain reactions based on a genome scale metabolic model. With comprehensive examples, for different species and environmental contexts, we show the importance and scope of applications and highlight the improvement compared to common boundary formulations in existing metabolic models. Our method allows for the contextualization of dynamic metabolic models based on all available information. We anticipate this to greatly increase their accuracy and predictive power for basic research and also for drug development and industrial applications.
- This article is part of the themed collection: Integrative Approaches for Signalling and Metabolic Networks

 Please wait while we load your content...
Please wait while we load your content...