Geometrical alignment for improving cell evaluation in a microchannel with application on multiple myeloma red blood cells
Abstract
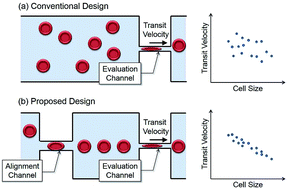
A microfluidic design for evaluating red blood cell deformability with geometrical alignment mechanism is proposed. While the transit velocity of a cell passing through a constriction channel is conventionally utilized as an index of cell deformability, the flow-in angle of the cell entering the channel is found affecting the transit significantly, and would interfere with the evaluation. To suppress such kind of interference, an additional alignment channel is placed in front of the evaluation channel for aligning target cells before entering the evaluation channel. Cells are spontaneously aligned by the geometrical constraints without any additional control. The experiments on the red blood cells from three healthy subjects and a patient with multiple myeloma are conducted. According to the Experimental results, the alignment channel effectively reduce 42.7% of position distribution in average, and the negative correlation between transit velocity and cell size is increased. The correlation shows the improved size sensitivity of the proposed method. For the purpose of comparison and validating the microchannel approach, the stiffness of the subjects' red blood cells is also measured by an atomic force microscope, the current gold standard of cell stiffness measurement. The results from two approaches show the same tendency of the RBC deformability, which evidently support the validity of the proposed method on cell deformability evaluation.

 Please wait while we load your content...
Please wait while we load your content...