Evaluation of a focused virtual library of heterobifunctional ligands for Clostridium difficile toxins†
Abstract
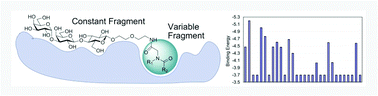
A focused library of virtual heterobifunctional ligands was generated in silico and a set of ligands with recombined fragments was synthesized and evaluated for binding to Clostridium difficile toxins. The position of the trisaccharide fragment was used as a reference for filtering docked poses during virtual screening to match the trisaccharide ligand in a crystal structure. The peptoid, a diversity fragment probing the protein surface area adjacent to a known binding site, was generated by a multi-component Ugi reaction. Our approach combines modular fragment-based design with in silico screening of synthetically feasible compounds and lays the groundwork for future efforts in development of composite bifunctional ligands for large clostridial toxins.

 Please wait while we load your content...
Please wait while we load your content...