Modelling ligand selectivity of serine proteases using integrative proteochemometric approaches improves model performance and allows the multi-target dependent interpretation of features†
Abstract
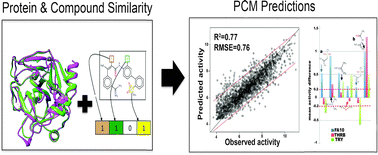
Serine proteases, implicated in important physiological functions, have a high intra-family similarity, which leads to unwanted off-target effects of inhibitors with insufficient selectivity. However, the availability of sequence and structure data has now made it possible to develop approaches to design pharmacological agents that can discriminate successfully between their related binding sites. In this study, we have quantified the relationship between 12 625 distinct protease inhibitors and their bioactivity against 67 targets of the serine protease family (20 213 data points) in an integrative manner, using proteochemometric modelling (PCM). The benchmarking of 21 different target descriptors motivated the usage of specific binding pocket amino acid descriptors, which helped in the identification of active site residues and selective compound chemotypes affecting compound affinity and selectivity. PCM models performed better than alternative approaches (models trained using exclusively compound descriptors on all available data, QSAR) employed for comparison with R2/RMSE values of 0.64 ± 0.23/0.66 ± 0.20 vs. 0.35 ± 0.27/1.05 ± 0.27 log units, respectively. Moreover, the interpretation of the PCM model singled out various chemical substructures responsible for bioactivity and selectivity towards particular proteases (thrombin, trypsin and coagulation factor 10) in agreement with the literature. For instance, absence of a tertiary sulphonamide was identified to be responsible for decreased selective activity (by on average 0.27 ± 0.65 pChEMBL units) on FA10. Among the binding pocket residues, the amino acids (arginine, leucine and tyrosine) at positions 35, 39, 60, 93, 140 and 207 were observed as key contributing residues for selective affinity on these three targets.
- This article is part of the themed collection: Computational Integrative biology (IB)


 Please wait while we load your content...
Please wait while we load your content...