Feasibility of occurrence of different types of protonated base pairs in RNA: a quantum chemical study†
Abstract
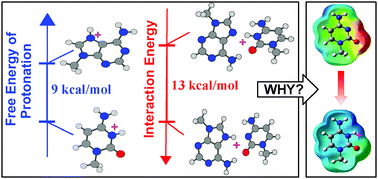
Protonated nucleobases have significant roles in facilitating catalytic functions of RNA, and in stabilizing different structural motifs. Reported pKa values of nucleobase protonation suggest that the population of neutral nucleobases is 103–104 times higher than that of protonated nucleobases under physiological conditions (pH ∼ 7.4). Therefore, a molecular level understanding of various putative roles of protonated nucleobases cannot be achieved without addressing the question of how their occurrence propensities and stabilities are related to the free energy costs associated with the process of protonation under physiological conditions. With water as the proton donor, we use advanced QM methods to evaluate the site specific protonation propensities of nucleobases in terms of their associated free energy changes (ΔGprot). Quantitative follow up on the energetics of base pair formation and database search for evaluating their occurrence frequencies, reveal a lack of correlation between base pair stability and occurrence propensities on the one hand, and ease of protonation on the other. For example, although N7 protonated adenine (ΔGprot = 40.0 kcal mol−1) is found to participate in stable base pairing, base pairs involving N7 protonated guanine (ΔGprot = 36.8 kcal mol−1), on geometry optimization, converge to a minima where guanine transfers its extra proton to its partner base. Such observations, along with examples of weak base pairs involving N3 protonation of cytosine (ΔGprot = 37.0 kcal mol−1) are rationalized by analysing the protonation induced charge redistributions which are found to significantly influence, both positively and negatively, the hydrogen bonding potentials of different functional sites of individual nucleobases. Protonation induced charge redistribution is also found to strongly influence (i) the aromatic character of the rings of the participating bases and (ii) hydrogen bonding potential of the free edges of the protonated base pair. Comprehensive analysis of a non-redundant RNA crystal structure dataset further reveals that, while availability of stabilization possibilities determine the feasibility of occurrence of protonated bases, their occurrence context and specific functional roles are important factors determining their occurrence propensities.


 Please wait while we load your content...
Please wait while we load your content...