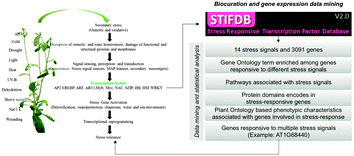
Biotic and abiotic stresses adversely affect agriculture by reducing crop growth and productivity worldwide. To investigate the abiotic stress-responsive genes in Arabidopsis thaliana, we compiled a dataset of stress signals and differentially upregulated genes (>= 2.5 fold change) from Stress-responsive transcription Factors DataBase (STIFDB) with additional set of stress signals and genes curated from PubMed and Gene Expression Omnibus. A dataset of 3091 genes differentially upregulated due to 14 different stress signals (abscisic acid, aluminum, cold, cold–drought–salt, dehydration, drought, heat, iron, light, NaCl, osmotic stress, oxidative stress, UV-B and wounding) were curated and used for the analysis. Details about stress-responsive enriched genes and their association with stress signals can be obtained from STIFDB2 database http://caps.ncbs.res.in/stifdb2. The gene–stress-signal data were analyzed using an enrichment-based meta-analysis framework consisting of two different ontologies (Gene Ontology and Plant Ontology), biological pathway and functional domain annotations. We found several shared and distinct biological processes, cellular components and molecular functions associated with stress-responsive genes. Pathway analysis revealed that stress-responsive genes perturbed the pathways under the “Metabolic pathways” category. We also found several shared and stress-signal specific protein domains, suggesting functional mechanisms regulating stress-response. Phenomic characteristics of abiotic stress-responsive genes were ascertained for several stresses and found to be shared by multiple stresses in both anatomy and temporal categories of Plant Ontology. We found several constitutive stress-responsive genes that are differentially upregulated due to perturbation of different stress signals, for example a gene (AT1G68440) involved in phenylpropanoid metabolism and polyamine catabolism as responsive to seven different stress signals. We also performed structure–function prediction of five genes associated responsive to multiple abiotic stress signals. We envisage that results from our analysis that provide insight into functional repertoire, metabolic pathways and phenomic characteristics common and specifically associated with stress signals would help to understand abiotic stress regulome in Arabidopsis thaliana and may also help to develop an improved plant variety using molecular breeding and genetic engineering techniques that are rapidly stress-responsive and tolerant.

 Please wait while we load your content...
Please wait while we load your content...