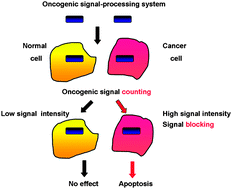
Synthesizing oncogenic signal-processing systems that function as both “signal counters” and “signal blockers” in cancer cells†
Abstract
RNA–
* Corresponding authors
a
Shenzhen Second People's Hospital, First Affiliated Hospital of Shenzhen University, Sungang Road, Shenzhen 518039, China
E-mail:
caizhiming2000@yahoo.com.cn
b Anhui Medical University, Hefei 230032, China
c
Guangdong and Shenzhen Key Laboratory of Male Reproductive Medicine and Genetics, Institute of Urology, Peking University Shenzhen Hospital, Shenzhen PKU-HKUST Medical Center, Lianhua Road, Shenzhen 518036, China
E-mail:
guiyaoting2007@yahoo.com.cn
RNA–

 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
Y. Liu, W. Huang, D. Zhou, Y. Han, Y. Duan, X. Zhang, H. Zhang, Z. Jiang, Y. Gui and Z. Cai, Mol. BioSyst., 2013, 9, 1909 DOI: 10.1039/C3MB70093C
To request permission to reproduce material from this article, please go to the Copyright Clearance Center request page.
If you are an author contributing to an RSC publication, you do not need to request permission provided correct acknowledgement is given.
If you are the author of this article, you do not need to request permission to reproduce figures and diagrams provided correct acknowledgement is given. If you want to reproduce the whole article in a third-party publication (excluding your thesis/dissertation for which permission is not required) please go to the Copyright Clearance Center request page.
Read more about how to correctly acknowledge RSC content.
 Fetching data from CrossRef.
Fetching data from CrossRef.
This may take some time to load.
Loading related content
