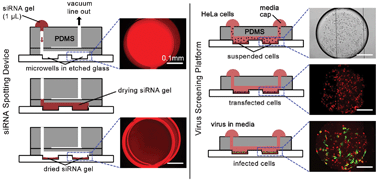
RNA interference (RNAi) is a powerful tool for functional genomics with the capacity to comprehensively analyze host–pathogen interactions. High-throughput RNAi screening is used to systematically perturb cellular pathways and discover therapeutic targets, but the method can be tedious and requires extensive capital equipment and expensive reagents. To aid in the development of an inexpensive miniaturized RNAi screening platform, we have developed a two part microfluidic system for patterning and screening gene targets on-chip to examine cellular pathways involved in virus entry and infection. First, a multilayer polydimethylsiloxane (PDMS)-based spotting device was used to array siRNA molecules into 96 microwells targeting markers of endocytosis, along with siRNA controls. By using a PDMS-based spotting device, we remove the need for a microarray printer necessary to perform previously described small scale (e.g. cellular microarrays) and microchip-based RNAi screening, while still minimizing reagent usage tenfold compared to conventional screening. Second, the siRNA spotted array was transferred to a reversibly sealed PDMS-based screening platform containing microchannels designed to enable efficient cell loading and transfection of mammalian cells while preventing cross-contamination between experimental conditions. Validation of the screening platform was examined using Vesicular stomatitis virus and emerging pathogen Rift Valley fever virus, which demonstrated virus entry pathways of clathrin-mediated endocytosis and caveolae-mediated endocytosis, respectively. The techniques here are adaptable to other well-characterized infection pathways with a potential for large scale screening in high containment biosafety laboratories.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...