Computational models for large-scale simulations of facilitated diffusion
Abstract
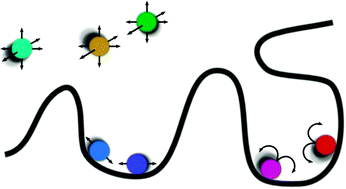
The binding of site-specific transcription factors to their genomic target sites is a key step in gene regulation. While the genome is huge, transcription factors belong to the least abundant
 Please wait while we load your content...
Please wait while we load your content...