Construction of large signaling pathways using an adaptive perturbation approach with phosphoproteomic data†
Abstract
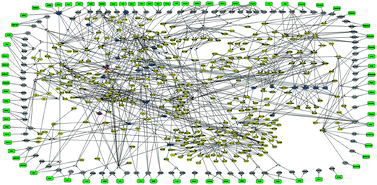
Construction of large and cell-specific signaling pathways is essential to understand information processing under normal and pathological conditions. On this front, gene-based approaches offer the advantage of large pathway exploration whereas phosphoproteomic approaches offer a more reliable view of pathway activities but are applicable to small pathway sizes. In this paper, we demonstrate an experimentally adaptive approach to construct large signaling pathways from phosphoproteomic data within a 3-day time frame. Our approach—taking advantage of the fast turnaround time of the xMAP technology—is carried out in four steps: (i) screen optimal pathway inducers, (ii) select the responsive ones, (iii) combine them in a combinatorial fashion to construct a phosphoproteomic dataset, and (iv) optimize a reduced generic pathway via an Integer Linear Programming formulation. As a case study, we uncover novel players and their corresponding pathways in primary human hepatocytes by interrogating the signal transduction downstream of 81 receptors of interest and constructing a detailed model for the responsive part of the network comprising 177 species (of which 14 are measured) and 365 interactions.

 Please wait while we load your content...
Please wait while we load your content...