Symmetric Epistasis Estimation (SEE) and its application to dissecting interaction map of Plasmodium falciparum†
Abstract
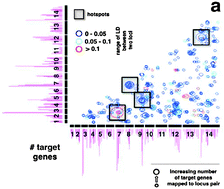
It is being increasingly recognized that many important phenotypic traits, including various diseases, are governed by a combination of weak genetic effects and their interactions. While the detection of epistatic interactions that involve a non-additive effect of two loci on a quantitative trait is particularly challenging, this interaction type is fundamental for the understanding of genome organization and gene regulation. However, current methods that detect epistatic interactions typically rely on the existence of a strong primary effect, considerably limiting the sensitivity of the search. To fill this gap, we developed a new method, SEE (Symmetric Epistasis Estimation), allowing the genome-wide detection of epistatic interactions without the need for a strong primary effect. We applied our approach to progeny crosses of the human malaria parasite P. falciparum and S. cerevisiae. We found an abundance of epistatic interactions in the parasite and a much smaller number of such interactions in yeast. The genome of P. falciparum also harboured several epistatic interaction hotspots that putatively play a role in

 Please wait while we load your content...
Please wait while we load your content...