An integrated image analysis platform to quantify signal transduction in single cells†
Abstract
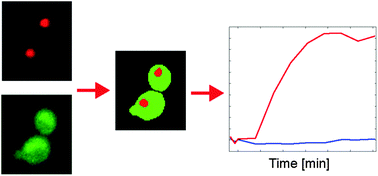
* Corresponding authors
a
ETH-Zurich, Institute of Biochemistry, Schafmattstr. 18, CH-8093 Zurich, Switzerland
E-mail:
serge.pelet@bc.biol.ethz.ch, matthias.peter@bc.biol.ethz.ch

 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
 Fetching data from CrossRef.
Fetching data from CrossRef.
This may take some time to load.
Loading related content
