Assigning kinetic 3D-signatures to glycocodes†
Abstract
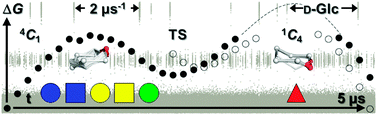
Reconciling glycocodes and their associated bioactivities, via 3D-structure, will rationalise burgeoning high-throughput functional glycomics data and underpin a new era of opportunity in chemical biology. A major impasse to achieving this goal is a detailed understanding of pyranose sugar ring 3D-conformation (or pucker) and the affiliated microsecond-timescale exchange kinetics. Here, we perform hardware-accelerated kinetically-rigorous equilibrium simulations of fundamental

 Please wait while we load your content...
Please wait while we load your content...