Biochip data normalization using multifunctional probes
Abstract
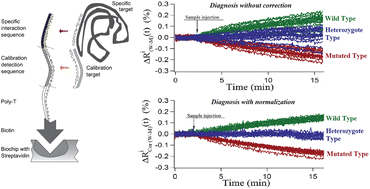
Using a biochip with stable probe functionalization and a detection system capable of real time measurements, it is demonstrated that acquired probe–target interaction data are more reproducible in time – on a given probe spot using sequential target runs – than in space, using many probe spot replicates on the biochip in one single parallel target run. To increase the biochip data precision, a normalization method that quantifies and corrects the surface inhomogeneity without the use of complex data post-processing has been developed. This simple and effective method is based on adding a common reactive group to all probes and quantifying the biochip response to a calibration target, thus quantifying the spatial heterogeneity in the biosensor responsiveness. The usefulness of such methodology, which can be easily generalized, is demonstrated in the model case of DNA:DNA interactions, using a surface plasmon resonance imaging system as the dynamical reader. The biochips are based on streptavidin biochemically functionalized gold films onto which biotinylated ssDNA probe sequences, related to cystic fibrosis genotyping, are spotted. This normalization method provides high gain in data precision and allows, in this example, unambiguous genotyping of SNP, including discrimination of the heterozygote case from the two homozygote cases.

 Please wait while we load your content...
Please wait while we load your content...