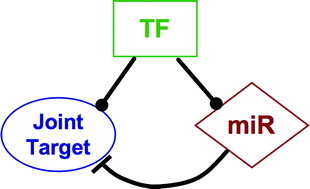
In this work, we describe a computational framework for the genome-wide identification and characterization of mixed transcriptional/post-transcriptional regulatory circuits in humans. We concentrated in particular on feed-forward loops (FFL), in which a master transcription factor regulates a microRNA, and together with it, a set of joint target protein coding genes. The circuits were assembled with a two step procedure. We first constructed separately the transcriptional and post-transcriptional components of the human regulatory network by looking for conserved over-represented motifs in human and mouse promoters, and 3′-UTRs. Then, we combined the two subnetworks looking for mixed feed-forward regulatory interactions, finding a total of 638 putative (merged) FFLs. In order to investigate their biological relevance, we filtered these circuits using three selection criteria: (I) GeneOntology enrichment among the joint targets of the FFL, (II) independent computational evidence for the regulatory interactions of the FFL, extracted from external databases, and (III) relevance of the FFL in cancer. Most of the selected FFLs seem to be involved in various aspects of organism development and differentiation. We finally discuss a few of the most interesting cases in detail.
This article is Open Access
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...