Identifying targets of miR-143 using a SILAC-based proteomic approach†
Abstract
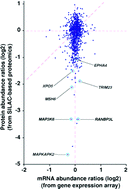
Although the targets of most miRNAs have not been experimentally identified, microRNAs (miRNAs) have begun to be extensively characterized in physiological, developmental and disease-related contexts in recent years. Thus far, mainly computational approaches have been employed to predict potential targets for the large majority of miRNAs. Although miRNAs exert a major influence on the efficiency of translation of their targets in animals, most studies describing experimental identification of miRNA target genes are based on detection of altered mRNA levels. miR-143 is a miRNA involved in tumorigenesis in multiple types of cancer, smooth muscle cell fate and adipocyte differentiation. Only a few miR-143 targets are experimentally verified, so we employed a SILAC-based quantitative proteomic strategy to systematically identify potential targets of miR-143. In total, we identified >1200 proteins from MiaPaCa2 pancreatic cancer cells, of which 93 proteins were downregulated >2-fold in miR-143 mimic transfected cells as compared to controls. Validation of 34 of these candidate targets in luciferase assays showed that 10 of them were likely direct targets of miR-143. Importantly, we also carried out gene expression profiling of the same cells and observed that the majority of the candidate targets identified by proteomics did not show a concomitant decrease in mRNA levels confirming that miRNAs affect the expression of most targets through translational inhibition. Our study clearly demonstrates that quantitative proteomic approaches are important and necessary for identifying miRNA targets.

 Please wait while we load your content...
Please wait while we load your content...