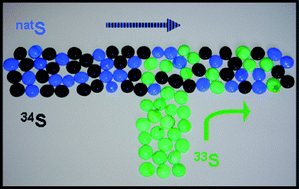
Different analytical strategies, such as Compound Independent Calibration (CIC), post-column Isotope Dilution Analysis (IDA) and post-column Isotope Pattern Deconvolution (IPD) were evaluated for the determination of sulfur-containing biomolecules. CIC graphs were obtained for the sulfur-containing compounds sulfate, cysteine, glutathione and methionine separated by isocratic HPLC using four different ICP-MS instruments. Instruments evaluated included two quadrupole based instruments (XSeries II and HP-4500), with and without a collision cell respectively, one single collector double focusing (Element 2) and one multicollector double focusing (Neptune). When isocratic HPLC separations were employed, CIC using sulfate standards could be applied for the determination of small peptides and amino acids in their native form. When gradient elution HPLC separations were performed, CIC was no longer suitable and enriched stable isotopes (33S) were evaluated to compensate for gradient-derived changes in the sensitivity for sulfur in the ion source. It was observed that post-column IDA using 33S was suitable for the absolute quantification of sulfate, cysteine, glutathione and methionine by HPLC-ICP-MS when gradient elution was performed. Unfortunately, traditional equations used for post-column IDA can no longer be applied when two different sulfur enriched isotopes are employed. In the characterisation of 34S-labelled yeast we will have three different sulfur isotope patterns: natural abundance sulfur impurities, 34S used as “labelling” tracer and 33S used as post-column “quantification” tracer. So, for the characterisation of 34S-labelled yeast we have developed a post-column IPD procedure that allowed us to obtain the “pattern specific molar flow chromatograms” and, hence, to discriminate between “natural abundance” and “34S-enriched” sulfur species in the 34S-labelled yeast.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...