The topology of drug–target interaction networks: implicit dependence on drug properties and target families†‡
Abstract
The availability of interaction data between small molecule
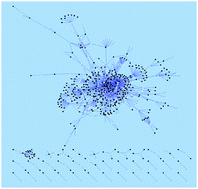
- This article is part of the themed collection: Emerging investigators

 Please wait while we load your content...
Please wait while we load your content...