Elongation and migration of single DNA molecules in microchannels using oscillatory shear flows†
Abstract
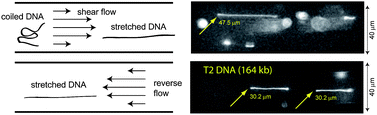
Much of modern biology relies on the strategic manipulation of molecules for creating ordered arrays prior to high throughput molecular analysis. Normally, DNA arrays involve deposition on surfaces, or confinement in nanochannels; however, we show that microfluidic devices can present stretched molecules within a controlled flow in ways complementing surface modalities, or extreme confinement conditions. Here we utilize pressure-driven oscillatory shear flows generated in microchannels as a new way of stretching DNA molecules for imaging “arrays” of individual DNA molecules. Fluid shear effects both stretch DNA molecules and cause them to migrate away from the walls becoming focused in the centerline of a channel. We show experimental findings confirming simulations using Brownian dynamics accounting for hydrodynamic interactions between molecules and channel-flow boundary conditions. Our findings characterize DNA elongation and migration phenomena as a function of molecular size, shear rate, oscillatory frequency with comparisons to computer simulation studies.

 Please wait while we load your content...
Please wait while we load your content...