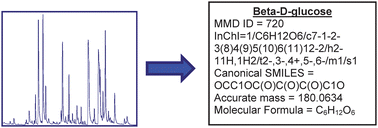
The chemical identification of mass spectrometric signals in metabolomic applications is important to provide conversion of analytical data to biological knowledge about metabolic pathways. The complexity of electrospray mass spectrometric data acquired from a range of samples (serum, urine, yeast intracellular extracts, yeast metabolic footprints, placental tissue metabolic footprints) has been investigated and has defined the frequency of different ion types routinely detected. Although some ion types were expected (protonated and deprotonated peaks, isotope peaks, multiply charged peaks) others were not expected (sodium formate adduct ions). In parallel, the Manchester Metabolomics Database (MMD) has been constructed with data from genome scale metabolic reconstructions, HMDB, KEGG, Lipid Maps, BioCyc and DrugBank to provide knowledge on 42,687 endogenous and exogenous metabolite species. The combination of accurate mass data for a large collection of metabolites, theoretical isotope abundance data and knowledge of the different ion types detected provided a greater number of electrospray mass spectrometric signals which were putatively identified and with greater confidence in the samples studied. To provide definitive identification metabolite-specific mass spectral libraries for UPLC-MS and GC-MS have been constructed for 1,065 commercially available authentic standards. The MMD data are available at http://dbkgroup.org/MMD/
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...