How well can new-generation density functional methods describe stacking interactions in biological systems?†
Abstract
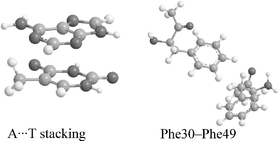
We compare the performance of four recently developed DFT methods (MPW1B95, MPWB1K, PW6B95, and PWB6K) and two previous, generally successful DFT methods (B3LYP and B97-1) for the calculation of stacking interactions in six nucleic acid bases complexes and five

 Please wait while we load your content...
Please wait while we load your content...