Open Access Article
Open Access ArticleCoordination chemistry of pyrazine derivatives analogues of PZA: design, synthesis, characterization and biological activity†
Małgorzata
Ogryzek
a,
Agnieszka
Chylewska
*a,
Aleksandra
Królicka
b,
Rafał
Banasiuk
b,
Katarzyna
Turecka
c,
Dorota
Lesiak
d,
Dawid
Nidzworski
de and
Mariusz
Makowski
*a
aLaboratory of Intermolecular Interactions, Faculty of Chemistry, University of Gdansk, W. Stwosza 63, 80-308 Gdansk, Poland. E-mail: agnieszka.chylewska@ug.edu.pl; mariusz.makowski@ug.edu.pl; Fax: +48 523 50 12; Tel: +48 58 523 50 55
bLaboratory of Biologically Active Compounds, Intercollegiate Faculty of Biotechnology, University of Gdansk and Medical University of Gdansk, Abrahama 58, 80-307 Gdansk, Poland
cFaculty of Pharmacy with Subfaculty of Laboratory Medicine, Al. Hallera 107, 80-416 Gdansk, Poland
dLaboratory of Molecular Virology, Intercollegiate Faculty of Biotechnology, University of Gdansk and Medical University of Gdansk, Kladki 24, 80-822 Gdansk, Poland
eInstitute of Biotechnology and Molecular Medicine, Trzy Lipy 3 St., 80-172 Gdańsk, Poland
First published on 23rd May 2016
Abstract
Ru(III) complexes with pyrazine derivatives: 2,3-bis(2-pyridyl)pyrazine (DPP), pyrazine-2-amidoxime (PAOX), pyrazine-2-thiocarboxamide (PTCA) and 2-amino-5-bromo-3-(methylamino)pyrazine (ABMAP) have been prepared. Characterization of the compounds was acquired using UV-Vis and FT-IR spectroscopy, elemental analysis, conductivity and electrochemical measurements as well as thermogravimetric studies. The ligand field parameters, Δ0 (splitting parameter), B (Racah parameter of interelectronic repulsion) and β (nephelauxetic ratio) were calculated. The stabilities of the Ru(III) complexes have also been confirmed by spectrophotometric titration methods in acetonitrile and water solutions. The data showed that the examined compounds are stable both in solution and solid states, which was also confirmed by the values of their stability constants found. Moreover, the molecular structures of the complexes have been optimized using AM1 and PM3 methods and this supported octahedral geometry around the Ru(III) ion. The minimum inhibitory (MIC) and minimum bactericidal concentration (MBC) for synthesized complexes were studied against two Gram (+) bacteria, two Gram (−) bacteria and fungi – three reference strains of Candida albicans. The results show that [RuCl(PAOX)2(OH2)]Cl2 display antifungal activity.
Introduction
In the past, transition metals and medicinal applications were thought to be mutually exclusive. Today, the therapeutic applications of transition metal complexes is an underdeveloped area of research. These complexes offer a great diversity in their application. Not only do they have anti-cancer properties but they have also been used as anti-inflammatory, anti-diabetic, anti-bacterial and anti-fungal compounds.1–3 The development of transition metal complexes as drugs is not an easy task; considerable effort is required to get a compound of interest. Besides all these limitations and side effects, coordination compounds are still the most widely used chemotherapeutics in a way that was unimaginable a few years ago.The number of platinum complexes that show antitumor activity is still rapidly growing, because of attempts to find complexes with their greater therapeutic potency and lower toxicity than existing clinical drugs. As a consequence, the attention has turned to other platinum group metals, like ruthenium, osmium, iridium and rhodium. A special attention has been focused on ruthenium compounds because they exhibit cytotoxicity against cancer cells and no cross-resistance with cis-platin.4 Ruthenium complexes demonstrate similar ligand exchange kinetics to those of platinum(II) antitumor drugs already used in clinical treatment, while displaying only low toxicity.5 This is partially due to the ability of ruthenium complexes to mimic the binding of iron to molecules of biological significance, exploiting mechanisms the organism has evolved for iron transport.6–8 The most promising metal-based anticancer drug candidates in clinical trials are the Ru(III) containing imidazolium trans-[tetrachlorido(1H-imidazole) (dimethylsulfoxide)ruthenate(III)] (NAMI-A) and trans-[tetrachloridobis(1H-indazole)ruthenate(III)] complexes (KP1019).9,10 KP1019 is efficient in colorectal carcinoma models, while NAMI-A is an antimetastatic agent which can affect the motility of cancer cells.11,12 The mode of action and the intracellular targets of Ru(III) complexes are not exactly known. There are many investigations suggesting that the intravenously injected drugs can be transported mainly by the serum albumin and/or transferrin in the blood plasma.13–16 The reduction of Ru(III) compounds in cytosol leads to the kinetically more labile and more reactive Ru(II) compounds. This is a result of the reductive atmosphere in tumor cells due to fast anabolic processes.9,17 DNA and cellular proteins like kinases or other enzymes were suggested as intracellular targets.18,19
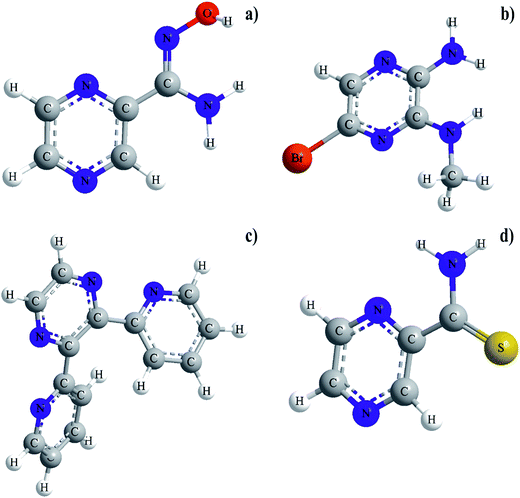
Our careful literature search indicated that ruthenium(III) complexes with pyrazinamide (PZA) analogue in which the amide moiety in a second position of the pyrazine ring has been replaced by thiocarboxamide group (PTCA), amidoxime moiety (PAOX), 2-pyridyl ring (DPP) and amine group (ABMAP), respectively, (Fig. 1) have not been very well recognized and their physicochemical and biological properties have been insufficiently studied.
Due to the low number of scientific publications and information on analogues of PZA, it is believed that the results of our research shown in this paper will be an original and significant contribution to the development of the field.
It is known that pyrazine ring is a part of polycyclic derivative and plays important biological and industrial roles.20 The pharmacological activities of the pyrazine derivatives vary and include substances with multidirectional actions. A low toxicity of these groups of compounds allows us to use them as a pharmacophore in designing new compounds to be used as drugs. The discovery of natural pyrazine derivatives, that showed the pharmacological effect, initiated the search for novel and more effective synthetic compounds exhibiting biological activities. There is a number of substances having antituberculotic21,22 and antibacterial activities,23,24 antifungal and cytotoxic effects,25,26 respectively, in the group of synthetic pyrazine derivatives. In addition, the compounds belonging to this group display antioxidant,27 antiproliferative,28 and antitumor activities.29
The compounds containing S, N and N, O donor atoms are important owing to their significant antifungal, antibacterial and anticancer activities.30,31 Cytotoxicity can be further improved by using ligands with O, N or N, N donor systems. A type of chelating ligands does not only have an influence on the biological properties but also bears impact on the stability of the formed complex.32–34 Furthermore, the ligand can modify the interaction with different biomolecules such as albumin, transferrin or various cellular proteins. It is well known that some drugs have increased their activities when administered as metal complexes rather than administered as free organic compounds.35–38 A large number of reports are available on the chemistry and biological activity of transition metal complexes containing O, N and S, N donor atoms, but reports on Ru(III) coordination compounds are limited.39,40
As a part of our studies about simple inorganic models of interest for the development of the bioinorganic chemistry of ruthenium, we have started the investigations on Ru(III) complexes with PAOX, PTCA, DPP and ABMAP. The selected analogues of PZA appear to be suitable models due to the preference of ruthenium for oxygen and nitrogen donor atoms in biological systems.
Recently, our research group have initiated a series of studies on the effect of different substituents at the second position of the pyrazine ring on the stereochemistry of the complexes formed. We observed the direct relation between the structure of a selected organic derivative as ligand and biological activity of the obtained coordination compounds. In this work, the chelation mechanism of selected pyrazine derivatives with Ru(III) was studied in order to show and understand the biosynthetic role of ruthenium(III) ion in vivo as well as to develop its bioactive compounds. The stabilities of Ru(III) complexes in aqueous solution have never been investigated using the spectroscopic method. Herein, the results of the first spectrophotometric determination of the gradual equilibrium constants of PAOX, PTCA, DPP and ABMAP with Ru(III) in acetonitrile (MeCN) are presented. The obtained data clearly demonstrated the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 metal to ligand complexes for all investigated compounds. Additionally, to the best of our knowledge, pyrazine-2-amidoxime, pyrazine-2-thiocarboxamide, and 2-amino-5-bromo-3-(methylamino)pyrazine have never been employed as ligands in transition metal complexes. Here, new ruthenium(III) chloride coordination compounds with pyrazine derivatives, together with the preliminary studies of their biological properties, are shown and described. It was shown in our previous studies that structures of the ligands had an important impact on the stability and the ability to inhibit microbial proliferation of compounds used.41–44 We believe the results of our studies will be helpful to further understand the binding mechanisms and can provide very important information for designing a new type of highly effective antifungal drugs. The obtained Ru(III) complexes are considered to be potential prodrugs; the knowledge of their speciation and the most plausible chemical forms in aqueous solution in the biologically relevant pH range is a mandatory prerequisite for understanding the alternations in their biological activity.
2 metal to ligand complexes for all investigated compounds. Additionally, to the best of our knowledge, pyrazine-2-amidoxime, pyrazine-2-thiocarboxamide, and 2-amino-5-bromo-3-(methylamino)pyrazine have never been employed as ligands in transition metal complexes. Here, new ruthenium(III) chloride coordination compounds with pyrazine derivatives, together with the preliminary studies of their biological properties, are shown and described. It was shown in our previous studies that structures of the ligands had an important impact on the stability and the ability to inhibit microbial proliferation of compounds used.41–44 We believe the results of our studies will be helpful to further understand the binding mechanisms and can provide very important information for designing a new type of highly effective antifungal drugs. The obtained Ru(III) complexes are considered to be potential prodrugs; the knowledge of their speciation and the most plausible chemical forms in aqueous solution in the biologically relevant pH range is a mandatory prerequisite for understanding the alternations in their biological activity.
The stability of Ru(III) complexes with selected pyrazine derivatives has never been investigated in aqueous solution with the use of spectroscopic method. Herein, the results of the first spectrophotometric determination of the gradual equilibrium constants of PAOX, PTCA, DPP and ABMAP with Ru(III) in acetonitrile (MeCN) and in aqueous solution are presented. A several researchers investigated stability of similar Ru(III) complexes, but they used solvolysis or hydrolysis of synthesized coordination compounds and they measured kinetic parameters of reactions.45–49 The main goal of our present studies is to find and explain the influence of metal ion on spectral properties of selected pyrazine derivatives. We observed the complex formation of defined stoichiometry (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) during addition small amount of metal ion to ligand solution and recorded spectral changes. This spectral changes, i.e. increase/decrease of intensity, appearance of isosbestic points, and batho- or hypsochromic effects are the results of interactions between the hard acid Ru(III) ion and donor atoms of ligand. Based on the results from spectrophotometric measurements the values of gradual and cumulative formation constants for Ru(III) complexes were determined. To our knowledge, the stability of Ru(III) complexes in both aqueous and MeCN solutions has never been investigated by using spectrophotometric procedure presented in this paper. Consequently, here we report the results of the first spectrophotometric determination of the gradual equilibrium constants of the ruthenium(III) complexes with analogues of PZA.
2) during addition small amount of metal ion to ligand solution and recorded spectral changes. This spectral changes, i.e. increase/decrease of intensity, appearance of isosbestic points, and batho- or hypsochromic effects are the results of interactions between the hard acid Ru(III) ion and donor atoms of ligand. Based on the results from spectrophotometric measurements the values of gradual and cumulative formation constants for Ru(III) complexes were determined. To our knowledge, the stability of Ru(III) complexes in both aqueous and MeCN solutions has never been investigated by using spectrophotometric procedure presented in this paper. Consequently, here we report the results of the first spectrophotometric determination of the gradual equilibrium constants of the ruthenium(III) complexes with analogues of PZA.
The main purpose of the present work is fully characterization of Ru(III) complexes with selected analogues of pyrazinamide in the solid state and in solution and the preliminary studies of their biological properties. Moreover, we focused also on the determination of the influence of the structure ligand on the stability and the ability to inhibit microbial proliferation of compounds used. We believe the results of our studies will be helpful to further understand the binding mechanisms and can provide very important information for designing a new type of highly effective antifungal drugs. The obtained Ru(III) complexes are considered to be potential prodrugs; the knowledge of their speciation and the most plausible chemical forms in aqueous solution in the biologically relevant pH range is a mandatory prerequisite for understanding the alternations in their biological activity.
Results and discussion
The synthesized Ru(III) complexes are non-hygroscopic (stable at room temperature) and in the form of amorphous solids but, unfortunately, crystals suitable for X-ray measurements were not available. Melting points of the newly synthesized ruthenium(III) coordination compounds were in the range of 256–280 °C. The complexes were soluble in water and in common organic solvents like acetonitrile and dimethyl sulfoxide. The results of elemental analysis and analytical data of the examined coordination compounds suggest that the metal to ligand ratio of the complexes is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry of the type [RuCl(py)2(OH2)]Cl for PTCA and PAOX and [RuCl2(py)2]Cl for ABMAP, 1
2 stoichiometry of the type [RuCl(py)2(OH2)]Cl for PTCA and PAOX and [RuCl2(py)2]Cl for ABMAP, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry of the type [RuCl(py)(OH2)3]Cl for DPP, respectively, where py stands for bidentate pyrazine derivatives. Selected physicochemical data are given in Table 1.
1 stoichiometry of the type [RuCl(py)(OH2)3]Cl for DPP, respectively, where py stands for bidentate pyrazine derivatives. Selected physicochemical data are given in Table 1.
| Complex | Colour | M. wt. (g mol−1) | Melting point (°C) | Yield (%) | % Found (calcd.) | |||
|---|---|---|---|---|---|---|---|---|
| C | H | N | S | |||||
| [RuCl(PTCA)2(OH2)]Cl2 (C10H12Cl3N6OS2Ru) | Dark brown | 503.80 | 275 | 96 | 23.84 (23.80) | 2.40 (2.45) | 16.68 (16.65) | 12.73 (12.70) |
| [RuCl(DPP)(OH2)3]Cl2 (C14H16Cl3N4O3Ru) | Black | 495.70 | 280 | 88 | 33.90 (33.92) | 3.23 (3.23) | 11.28 (11.28) | — |
| [RuCl2(ABMAP)2]Cl (C10H14Br2Cl3N8Ru) | Black green | 613.50 | 262 | 83 | 19.55 (19.58) | 2.26 (2.30) | 18.23 (18.26) | — |
| [RuCl(PAOX)2(OH2)]Cl2 (C10H14Cl3N8O3Ru) | Navy blue | 501.70 | 256 | 79 | 23.94 (23.90) | 2.81 (2.79) | 22.33 (22.31) | — |
The presence of water molecules in the compounds is deduced from elemental analysis, IR spectra and thermal analysis (TG). The presence of chloride counter ion in the Ru(III) complexes was detected with a few drops of the concentrated silver nitrate reagent and the appearance of the white precipitate.
Infrared spectra studies
The infrared absorption bands are one of the most important tools of analysis used to determine the mode of coordination. The IR spectra of free ligands were compared with the spectra of their Ru(III) complexes. Strong bands in the range of 3403–3437 cm−1 region in the spectra of coordination compounds studied with PTCA, PAOX and DPP were assigned to a ν(O–H) stretching and suggested the presence of water molecules in the coordination sphere of these compounds. New bands (Table 2) were observed at the far IR region between 646 and 471 cm−1 for all chloride Ru(III) complexes with pyrazine derivatives, which were absent in the spectrum of free ligands. These bands have been ascribed to prominent peak of stretching frequencies of ν(Ru–N) and ν(Ru–Cl) vibrations, respectively.| Complex | ν(OH) | ν(N–H) |
ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) N) |
ν(C–N) | ν(Ru–N) | ν(Ru–Cl) |
|---|---|---|---|---|---|---|
| [RuCl(PAOX)2(OH2)]Cl2 | 3403 | 3160 | 1642 | 1036 | 646 | 524 |
| [Ru(PTCA)2(OH2)2]Cl3·3H2O | 3419 | 3065 | 1579 | 1039 | 580 | 471 |
| [Ru(ABMAP)3]Cl3 | — | 3403 | 1550 | 1347 | 589 | 479 |
| [RuCl(DPP) (OH2)4]Cl2·7H2O | 3437 | — | 1634 | 1391 | 586 | 494 |
In the case of [RuCl(PAOX)2(OH2)]Cl2, a band at 3160 cm−1 in the spectrum of the complex can be attributed to the stretching vibration of NH2 moiety (see Fig. S1 of the ESI†). Moreover, the characteristic absorption at 1659 cm−1 in the free pyrazine-2-amidoxime can be assigned to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) stretching vibrations of azomethine nitrogen.50,51 After complexation, this frequency was observed to be shifted to lower wave numbers (1642 cm−1), which confirmed the involvement of a nitrogen atom in bonding with ruthenium(III) ion. The infrared spectrum of free PAOX showed a sharp band at 953 cm−1 due to ν(N–O) stretching vibrations of oxime group. The bathochromic effect of frequency (844 cm−1) was observed after synthesis, implying the possibility of coordination by a nitrogen atom of this moiety.
N) stretching vibrations of azomethine nitrogen.50,51 After complexation, this frequency was observed to be shifted to lower wave numbers (1642 cm−1), which confirmed the involvement of a nitrogen atom in bonding with ruthenium(III) ion. The infrared spectrum of free PAOX showed a sharp band at 953 cm−1 due to ν(N–O) stretching vibrations of oxime group. The bathochromic effect of frequency (844 cm−1) was observed after synthesis, implying the possibility of coordination by a nitrogen atom of this moiety.
In the spectrum of pyrazine-2-thiocarboxamide (see Fig. S2 of the ESI†) the bands at 3341 cm−1, and 3239 cm−1 are ascribed to symmetric and antisymmetric stretching vibrations of NH2 group, respectively. In comparison with the spectrum of the Ru(III) complex, these bands disappeared, which confirmed the participation of a nitrogen atom in the complexation of metal ion. The strong and sharp band at 1607 cm−1 in the case of pyrazine-2-thiocarboxamide can be attributed to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) stretching vibrations. The significant bathochromic effect of this frequency after synthesis implied a possibility of complexation by the nitrogen of the azomethine group.
N) stretching vibrations. The significant bathochromic effect of this frequency after synthesis implied a possibility of complexation by the nitrogen of the azomethine group.
The characteristic absorption of the azomethine group appeared at 1653 cm−1 in the spectrum of 2,3-bis(2-pyridyl)pyrazine (Fig. S3 of the ESI†). This frequency was observed to be shifted to lower wave numbers (1634 cm−1) after complexation, which confirmed the involvement of a nitrogen atom in bonding with metal ion. The strong band at 1391 cm−1 is designated to ν(C–N) stretching modes of vibrations, which is consistent with the assignment for aromatic compounds.52 In the spectrum of [RuCl(DPP)(OH2)3]Cl2 all characteristic bands, which can be attributed to aromatic stretching vibrations in the region between 1500–1000 cm−1, were absent in our measurements, indicating that a nitrogen atom of pyridyl ring was involved in complexation of ruthenium(III) ion.
The strong, sharp band at 3438 cm−1 in the spectrum of 2-amino-5-bromo-3-(methylamino)pyrazine can be attributed to ν(NH2) stretching vibrations (Fig. S4 of the ESI†). The bathochromic shift of this frequency to 3403 cm−1 after synthesis implied the possibility of complexation by the nitrogen atom of the amine group. Two bands at 1586 cm−1 and 1383 cm−1 occurred in the spectrum of ABMAP, which can be ascribed to ν(N–H) and ν(C–N) stretching vibrations, respectively. In comparison with its Ru(III) complex, these bands were shifted to 1550 cm−1 and 1347 cm−1, respectively. This observation suggested that ABMAP coordinated Ru(III) ion by two nitrogen atoms of amine and methylamine groups, respectively.
The analysis of presented IR spectra of studied complexes in this work leads to a conclusion that selected pyrazine derivatives behave as bidentate ligands. In case of pyrazine-2-amidoxime complex, a ligand is coordinated to the metal ion by azomethine nitrogen and nitrogen of oxime groups. In the [RuCl(PTCA)2(OH2)]Cl2 the pyrazine-2-thiocarboxamide is bonded to the Ru(III) ion through the amine and azomethine nitrogen atoms, respectively. And the binding set of Ru(III) complex with 2,3-bis-(2-pyridyl)pyrazine includes nitrogen of the azomethine group and pyridyl ring. In the [RuCl2(ABMAP)2]Cl, the ruthenium(III) ion is coordinated by two nitrogen atoms of amine and methylamine groups, respectively. This means that there were more energetically favorable five-membered rings formed during the complexation process, which was consistent with indicated coordination sites.
Proton nuclear magnetic resonance spectra studies
Although Ru(III) complexes are paramagnetic, proton nuclear magnetic resonance spectroscopy can provide important structural information for such compounds.53,54 The 1H NMR spectral data of ruthenium(III) complexes with selected pyrazine derivatives were recorded in DMSO-d6 (Fig. S5–S8 of ESI†). The intensities of all resonance lines were determined. The assignment of the characteristic signals was performed by comparison with spectra of similar Ru(III) complexes.55 The presented 1H NMR spectra have several broad peaks, in accordance with the presence of paramagnetic Ru(III) metal ions.The following conclusions can be derived by comparing the spectra of ligands and their coordination compounds. The signal due to N–H protons appeared at 7.61 ppm in the case of [RuCl2(ABMAP)2]Cl and 8.14 ppm for [RuCl(PTCA)2(OH2)]Cl2. The downfield shifts of this signal in Ru(III) complexes occurred. This is correlated to the decrease of electron density and the deshielding of proton because of participation of amine and azomethine groups upon coordination which was observed in earlier report for Ru(III) complexes.56 Furthermore, in the [RuCl2(ABMAP)2]Cl, the disappearance of signal due to N–H proton in the third position of pyrazine ring indicated participation of this group in chelation process. In the case of PAOX signal correspond to N–H proton occurred at 5.25 ppm, which remains unchanged in their ruthenium(III) complex. This observation confirmed that amine group is not involved in complexation process of metal ion. The peaks observed and in the range 8.93–8.74 ppm and 7.30–7.03 ppm in the spectra of Ru(III) complexes are assigned to the presence of aromatic proton of pyrazine ring. The downfield shifts of peaks correspond to H6 proton of pyrazine ring and H2 proton of pyridyl ring is correlated to the decrease electron density caused by participation of nitrogen atoms of azomethine group and pyridyl ring. The position of protons correspond well with the proposed structure of Ru(III) complexes and were assigned in view of earlier reports.57,58
Electrospray ionization mass spectrometry analysis (ESI-MS)
Mass spectroscopy, which is mainly used in the analysis of biomolecules, has been increasingly applied as a powerful tool of structural characterization in coordination chemistry.59–61 The ESI-MS spectra of Ru(III) complexes dissolved in acetonitrile are comparison with simulated data and the obtained results are presented in Fig. S9–S12 of ESI.† The mass spectra of compounds studies were recorded in the positive mode and in the range of m/z = 50–800. The molecular peaks for the complexes of Ru(III) were observed at m/z = 501.9, 503.8, 495.9 and 612.8 for PAOX, PTCA, DPP and ABMAP, respectively which corresponding to the actual molecular weights of these complexes. For all Ru(III) compounds studies, experimental and simulated data are in good agreement, which well correspond with previous report for similar type of complexes.62 The presented results of ESI-MS studies of each Ru(III) complexes with pyrazine derivatives support the proposed structure of the coordination compounds.Electronic spectra of complexes synthesized
The electronic absorption spectra of Ru(III) complexes with selected pyrazine derivatives were recorded in acetonitrile in the range 250–700 nm (Fig. 2). UV-Vis spectral data for synthesized Ru(III) complexes were given in Table 3.| Complex | v 1 (2T2g → 2A1g, 2T1g) | v 2 (2T2g → 2Eg) | v 3 (π–π*, n–π*) | v 2/v1 | Δ 0 | B | β | C |
|---|---|---|---|---|---|---|---|---|
| a The ligand field parameters (Δ0, B, C and β) were calculated by data obtained from the experimental UV-Vis spectra of ruthenium(III) complexes and Tanabe–Sugano diagram for d5 octahedral geometry.54 | ||||||||
| [RuCl(PTCA)2(OH2)]Cl2 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 194 194 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 038 038 |
31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 746 746 |
1.25 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 261 261 |
638 | 0.66 | 2552 |
| [RuCl(DPP) (OH2)3]Cl2 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 657 657 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 276 276 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258 258 |
1.14 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 560 |
616 | 0.64 | 2464 |
| [RuCl2(ABMAP)2]Cl | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 048 048 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 509 509 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 787 787 |
1.29 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450 450 |
691 | 0.72 | 2764 |
| [RuCl(PAOX)2(OH2)]Cl2 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 202 202 |
26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 178 |
37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 174 174 |
1.31 | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 631 631 |
639 | 0.66 | 2556 |
The ground state of ruthenium(III) ion (t52g configuration) is 2T2g. The first excited doublet levels in order of increasing energy are 2A2g and 2T1g which arise from t42ge1g configuration.63,64 These Ru(III) complexes are low spin (t52g) with one unpaired electron; hence the bands observed can attributed to ligand field transitions for ruthenium(III) with t52g configuration. In the octahedral ligand field symmetry the spectrum of ruthenium(III) should show the spin-allowed d–d bands in the visible region.65 According to Tanabe–Sugano energy matrices66,67 for d5-octahedral geometry, the bands appearing in the range of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 657–20
657–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 202 and 21
202 and 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 276–26
276–26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 cm−1 were assigned to 2T2g → 2A1g or 2T1g and 2T2g → 2Eg, respectively. The bands of high intensity in the range 31
178 cm−1 were assigned to 2T2g → 2A1g or 2T1g and 2T2g → 2Eg, respectively. The bands of high intensity in the range 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 746–37
746–37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 174 cm−1 were also observed. These bands were assigned to π → π* and n → π* intra-ligand charge transfer. The ligand field parameters could be estimated by using the energies 2T2g → 2A1g and/or 2T1g, and 2T2g → 2Eg transitions, and the Tanabe–Sugano energy matrices for d5-octahedral geometry.
174 cm−1 were also observed. These bands were assigned to π → π* and n → π* intra-ligand charge transfer. The ligand field parameters could be estimated by using the energies 2T2g → 2A1g and/or 2T1g, and 2T2g → 2Eg transitions, and the Tanabe–Sugano energy matrices for d5-octahedral geometry.
The values of splitting parameter (Δ0) in the range 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560–31
560–31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 631 cm−1 were obtained. The values of Racah parameter of interelectronic repulsion (B) in the range 616–691 cm−1 and nephelauxetic parameter (β) in the range 0.64–0.72 were found (see Table 3). The ligand field parameters (Δ0, B, β) are close to those reported for similar octahedral ruthenium(III) complexes.68,69 The values of the splitting parameters placed the pyrazine derivatives in the middle range of spectrochemical series and provide reassurance that these ligands were coordinated to the Ru(III) ion through the nitrogen donor atoms. The B values for ruthenium(III) complexes (lower than that of free ion) indicate a considerable orbital overlap with a strong covalent metal–ligand bond.70 According to Jorgensen,65 a decrease in B-value is associated with the reduction in the nuclear charge on the cation and an increasing tendency to be reduced. It is apparent that the β parameter depends greatly on the electronegativity of the donor atoms and the ligand structure. The values of nephelauxetic parameter (β) are also less than one and show that selected ligands are middle nitrogen donor series.
631 cm−1 were obtained. The values of Racah parameter of interelectronic repulsion (B) in the range 616–691 cm−1 and nephelauxetic parameter (β) in the range 0.64–0.72 were found (see Table 3). The ligand field parameters (Δ0, B, β) are close to those reported for similar octahedral ruthenium(III) complexes.68,69 The values of the splitting parameters placed the pyrazine derivatives in the middle range of spectrochemical series and provide reassurance that these ligands were coordinated to the Ru(III) ion through the nitrogen donor atoms. The B values for ruthenium(III) complexes (lower than that of free ion) indicate a considerable orbital overlap with a strong covalent metal–ligand bond.70 According to Jorgensen,65 a decrease in B-value is associated with the reduction in the nuclear charge on the cation and an increasing tendency to be reduced. It is apparent that the β parameter depends greatly on the electronegativity of the donor atoms and the ligand structure. The values of nephelauxetic parameter (β) are also less than one and show that selected ligands are middle nitrogen donor series.
Conductance
The conductivity values of 10−3 M solutions of the synthesized complexes were carried out in water at 25 °C and presented in Table 4. All systems studied have the conductivity in the range 686.50–127.5 μS cm−1. These values indicate that Ru(III) complexes are electrolytes. The conductivities of 10−3 M solutions of NiCl2 and NaCl in water were also measured as 713.00 and 133.70 μS cm−1, respectively. These values are approximate to the conductivity values of synthesized complexes. It means that the examined coordination compounds are of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 electrolyte types as compared to NaCl and NiCl2 solutions, respectively. These results confirmed that one or two chloride anions were counter ions in the coordination spheres of the ion complexes studied in this work.71
2 electrolyte types as compared to NaCl and NiCl2 solutions, respectively. These results confirmed that one or two chloride anions were counter ions in the coordination spheres of the ion complexes studied in this work.71
| Compound | Conductance (μS·cm−1) |
|---|---|
| [RuCl(PAOX)2(OH2)]Cl2 | 686.5 |
| [RuCl(PTCA)2(OH2)]Cl2 | 611.10 |
| [RuCl(DPP)(OH2)3]Cl2 | 580.60 |
| [RuCl2(ABMAP)2]Cl | 127.50 |
| NaCl | 133.70 |
| NiCl2 | 713.00 |
Thermal analysis
Thermal analysis provides significant information about the stability and distribution of water molecules in the coordination sphere of metal complexes. In case of PTCA and PAOX complexes of the type [RuCl(py)2(OH2)]Cl and DPP of type [RuCl(py)(OH2)3]Cl, respectively, decomposition involved three steps. The TG curve of [RuCl2(ABMAP)2]Cl showed two main consecutive steps of mass loss. In the argon atmosphere the mass loss in the temperature range 125–180 °C corresponds to the loss of coordinated water molecule. The second step was due to the removal of volatilization of appropriate pyrazine derivative and hydrochloride molecules together with chlorine (Cl2) gas moiety fragment and it occurred in the temperature range 180–460 °C. At the last step, as the final product, stable metal oxides such as Ru2O3 (sometimes even RuO) were found above 750 °C as carbonaceous matter (Table 5). In all cases, the DTG study reveals that all the decomposition stages were exothermic in nature.| Complex | Temp. range | % mass loss observed (calc.) | Probable composition of expelled group/residue |
|---|---|---|---|
| [RuCl(PTCA)2(OH2)]Cl2 | 125–140 | 3.50 (3.57) | Loss of lattice water |
| 180–400 | 76.55 (76.62) | Loss of volatilization of two PTCA and HCl molecules together with chlorine (Cl2) gas moiety fragment | |
| >750 | 19.95 (19.81) | Residue mass as ruthenium oxides | |
| [RuCl(DPP)(OH2)3]Cl2 | 140–180 | 10.79 (10.89) | Loss of three lattice water |
| 240–440 | 68.28 (68.97) | Loss of volatilization of one DPP and HCl molecules together with chlorine (Cl2) gas moiety fragment | |
| >765 | 20.93 (20.14) | Residue mass as ruthenium oxides | |
| [RuCl(PAOX)2(OH2)]Cl2 | 125–150 | 3.51 (3.59) | Loss of lattice water |
| 190–460 | 76.30 (76.24) | Loss of volatilization of two PAOX and HCl molecules together with chlorine (Cl2) gas moiety fragment | |
| >760 | 20.19 (20.17) | Residue mass as ruthenium oxides | |
| [RuCl2(ABMAP)2]Cl | 260–600 | 83.12 (83.42) | Loss of volatilization of two ABMAP and HCl molecules together with chlorine (Cl2) gas moiety fragment |
| >780 | 16.88 (16.58) | Residue mass as ruthenium oxides |
Electrochemical results
Ru(III)/Ru(II) reduction potentials change with ligand environment and is thought to be very important for possible antitumor properties of a compound. The redox behavior of the ruthenium(III)–pyrazine derivative complexes was studied in acetonitrile solution at a platinum working electrode. Acetonitrile was selected as a solvent because of its well-known72,73 coordination ability with Ru(II) or Ru(III) centers. The relevant electrochemical data were summarized in Table 6 and voltammograms were shown in Fig. 3.| Voltammetric data | |||||
|---|---|---|---|---|---|
| Complex | Reduction, RuIII/RuII | ||||
| Potential [V] | Current [A] | ||||
| E pa | E pc | ΔEp | E 1/2 | I pa/Ipc | |
| a Reference electrode – SCE; E1/2 = 0.5(Epa + Epc) where Epa and Epc are anodic and cathodic peak potential, respectively; ΔEp = Epa − Epc; *Epa of [RuCl(PTCA)2(OH2)]Cl2 is considered due to the irreversible nature of the voltammogram.** Potentials of ABMAP ligand reduction. | |||||
| [RuCl(PAOX)2(OH2)]Cl2 | −1.003 | −1.252 | 0.249 | −1.127 | 0.357 |
| [RuCl(PTCA)2(OH2)]Cl2 | −0.856 | −1.285 | 0.429 | −1.071 | 0.333 |
| [RuCl2(ABMAP)2]Cl | −0.871 | −1.163 | 0.292 | −1.017 | 0.273 |
| −0.116** | −0.552** | 0.436** | −0.334** | 0.367** | |
| [RuCl(DPP) (OH2)3]Cl2 | −0.953 | −1.347 | 0.394 | −1.150 | 0.308 |
| Complex | Oxidation, RuIII/RuIV | ||||
|---|---|---|---|---|---|
| Potential [V] | Current [A] | ||||
| E pa | E pc | ΔEp | E 1/2 | |Ipc/Ipa| | |
| [RuCl(PAOX)2(OH2)]Cl2 | 1.376 | 0.874 | 0.502 | 1.125 | 0.325 |
| [RuCl(DPP) (OH2)3]Cl2 | 1.312 | 0.906 | 0.406 | 1.109 | 0.788 |
| [RuCl(PTCA)2(OH2)]Cl2 | 1.302* | — | — | — | — |
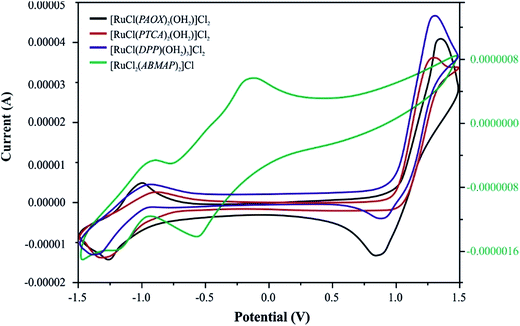 | ||
| Fig. 3 Cyclic voltammograms of four Ru(III) complexes with selected pyrazine derivatives (PAOX, PTCA, ABMAP, DPP) in acetonitrile solution (0.1 M TBAP) at a scan rate of 50 mV s−1. | ||
[RuCl(PAOX)2(OH2)]Cl2 and [RuCl(DPP)(OH2)3]Cl2 complexes exhibit two clear redox systems. On the anodic side reversible oxidations are observed and on the cathodic side, reversible redox couples. These redox systems can be assigned to the ruthenium(III)–ruthenium(IV) (positive potentials) and ruthenium(III)–ruthenium(II) (negative potentials) couples, respectively. Both the redox responses are reversible, as reflected in the equality of the anodic peak current (Ipa) with the cathodic peak current (Ipc). Though the peak-to-peak separation (ΔEp) is slightly larger (0.08 V) than ideally expected for a reversible electron transfer process.74 No changes in the ΔEp values were observed in relations to changes in the scan rate.
On the basis of the above-mentioned results, [RuCl(PTCA)2(OH2)]Cl2 and [RuCl2(ABMAP)2]Cl exhibit different behaviors. The reversible reduction process (RuIII → RuII, ΔEp = 0.429 V for complex with PTCA as ligand, and 0.292 V for complex with DPP) on cyclic voltammograms for these both complexes was observed. However, the nature of [RuCl(PTCA)2(OH2)]Cl2 voltammogram showed the irreversible process of oxidation in high, positive potentials. In the second case, for [RuCl2(ABMAP)2]Cl complex, cyclic voltammetry (CV) indicated additional reduction process in negative potentials, which could be ascribed to the reversible ligand reduction (ΔEp = 0.436 V).
Molecular structure and analysis of bonding modes
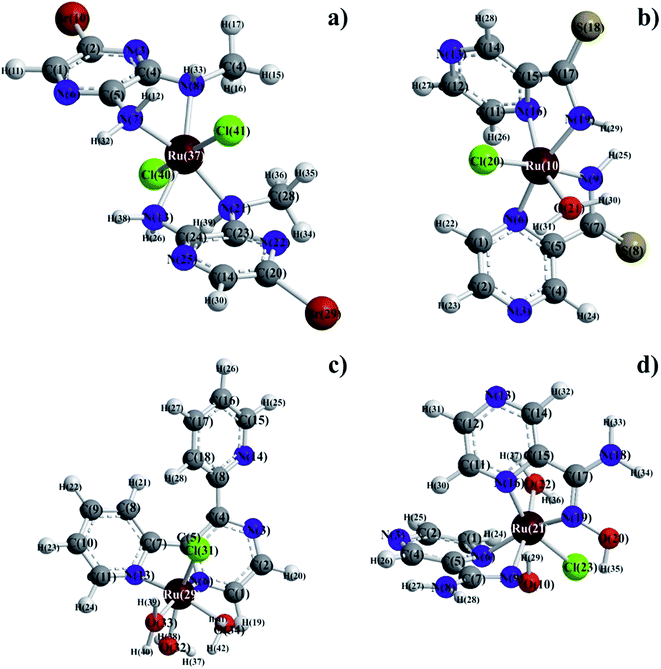
Molecular mechanics modeling was carried out to estimate the optimal electron energies, selected structure parameters, and to confirm the possibility of the studied compounds formation. In order to ascertain the structural preferences and coordination behavior of pyrazine derivatives (L) to ruthenium(III) ion, molecular mechanics calculations on the [RuLn]3+ species (1 < n < 3) were undertaken. Energy minimization was repeated several times to find the global minimum. The bond lengths and angles of all complexes were presented in Tables S1–S8,† respectively. The results of calculations have proven clear octahedral sphere of Ru(III) complexes and the formation of stable coordination compounds. It is important to note that in all four complexes Ru–O and Ru–N bond lengths are almost the same (1.89 < d [Å] < 1.95), the only difference being in bond angles. The analysis of Ru–Cl bond lengths showed similar values of about 2.25 Å for complexes of Ru(III) ions with ABMAP, PTCA and PAOX; except for complex cation with 2,3-bis(2-pyridyl)pyrazine – [RuCl(DPP)(OH2)3]+, where Ru–Cl bond length is shorter, i.e. 1.93 Å. The bond angles in all complexes are quite near to an octahedral arrangement predicting sp3d2 hybridization (Fig. 4 and S13†).The final coordinates and their heat of formations found by the PM3 method were shown in Table S9 of the ESI.†
Complexometric properties of pyrazine derivatives analogues of PZA
The presence of chromophore groups in a molecule of a ligand, whose absorption spectra change upon complexation, allows for using spectroscopy methods to study chelation process. Spectrophotometric titration is a very useful method to determine the stability constants in solution. Based on the results from spectroscopy measurements, it is possible to show a quantity of equilibria existing in a particular solution. Furthermore, this investigation can provide complete information about species formed during the titration process.The stability constants of pyrazine derivatives complexes with ruthenium(III) ion were determined by titration of the bidentate ligand (L–L) and recorded the spectral changes in the range 200–600 nm in MeCN (Fig. 5–8) and in aqueous solution (see Fig. S14–S17 of ESI†). Water was selected as a solvent because of its crucial role in microbiological assay. The change of the solvent from water to acetonitrile was made because of its better coordination ability with ruthenium(III) ion.
The values of stability constants for investigated complexes were calculated using EQUID computer program.75 This program is based on the non-linear least-square Gauss–Newton–Marquardt method for fitting procedure.75,76 The gradual (K) and cumulative (β) stability constants can be described by the following eqn (1)–(3):
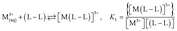 | (1) |
 | (2) |
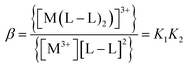 | (3) |
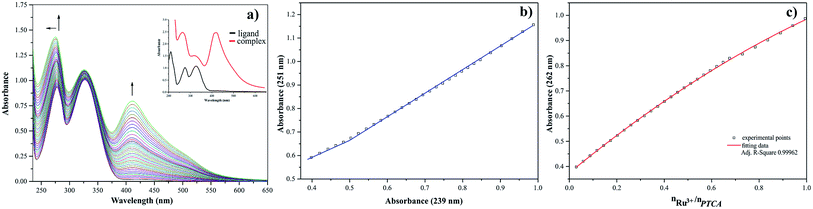
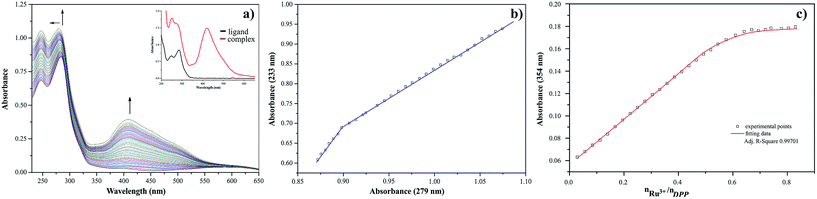
The presence of metal ion also influences spectral properties of the investigated ligands. The interaction of Ru(III) with pyrazine-2-thiocarboxamide induces changes in the absorption band positions after complexation. To determine the stability constants of PTCA with ruthenium(III), the ligand studied was titrated by acetonitrile solution of metal chloride salt and PTCA. Based on the analysis of the spectrophotometric titration curves, the number of equilibria and stoichiometry of formed complex were determined (Fig. 5a). The intensities of absorption bands increase and hyperchromic effect can be also observed. At the same time a new peak appeared at 412 nm. This observation suggests the existence of metal ion interactions with hard donor atom of PTCA in the system studied.
Dependence of the absorbance at 251 nm to the absorbance at 239 nm Ru(III)–PTCA system was obtained (see Fig. 5b). Two straight sections presented on the plot confirmed two equilibria during chelation process. The dependence of absorbance at 262 nm for 2-thiocarboxamide as a function of molar ratio nRu(III)/nPTCA is presented in Fig. 5c. The fitted data confirmed metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand stoichiometry 1
ligand stoichiometry 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 of the complex formation process.
2 of the complex formation process.
The results of spectrophotometric titration for Ru(III) ion with 2,3-bis(2-pyridyl)pyrazine system studied in MeCN solution have been shown in Fig. 6a. The intensities of absorption bands gradually increase and a slight hypsochromic shift can be observed. The changes in absorbance values are associated with chelation of nitrogen donor atom to ruthenium(III) cation. The A-diagrams (the dependence of absorbance at 233 nm as a function of absorbance at 279 nm) were plotted to illustrate specific quantity equilibria constants in arrangement studied (see Fig. 6b). The presence of two straight sections in these diagrams indicate two equilibria in the system studied. These observations are confirmed by the dependence of absorbance at 354 nm for 2,3-bis(2-pyridyl)pyrazine as a function of molar ratio nRu(III)/nDPP (see Fig. 6c). The metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand stoichiometry 1
ligand stoichiometry 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in arrangement of the studied complex is formed, which is consistent with values of molar ratio equal 0.5.
2 in arrangement of the studied complex is formed, which is consistent with values of molar ratio equal 0.5.
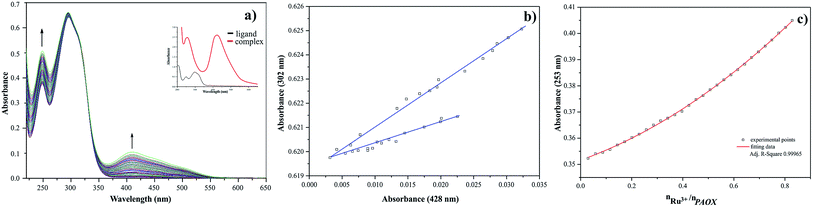
During the titration process of pyrazine-2-amidoxime by the ruthenium(III) chloride in acetonitrile solution with PAOX a slight shift to longer wavelengths (bathochromic effect) occurred. At the same time the maximum of band at 410 nm can be observed (Fig. 7a). These effects suggest the existence of metal ion interactions with donor atom of ligand in the system studied. The A-diagrams were analyzed to determine the quantity of equilibria present in the arrangement studied. Two straight sections in the dependence of the absorbance at 279 nm to the absorbance at 428 nm confirmed the existence of two equilibria during the chelation process (Fig. 7b). The major stoichiometry of complex formation is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, which corresponds to 0.5 value of molar ratio (Fig. 7c).
2, which corresponds to 0.5 value of molar ratio (Fig. 7c).
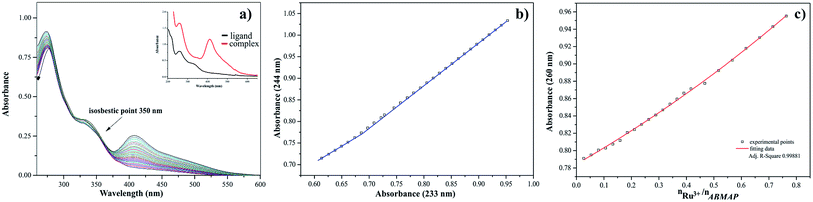
The results of titration process for ruthenium(III)-2-amino-5-bromo-3-(methylamino)pyrazine arrangement in MeCN have been presented in Fig. 8a. The intensities of absorption bands increase, indicating a small hypsochromic effect. At the same time the isosbestic point and new maximum of absorption appeared at 350 nm and 414 nm, respectively. The presented spectral changes are the results of interactions between the hard acid – Ru(III) ion and donor atoms of ligand. To define the accurate number of equilibria presented in the system studied, the A-diagrams were plotted. The dependence of the absorbance at 244 nm to the absorbance at 233 nm showed two straight sections (Fig. 8b), suggesting that two formation constants could be determined in this case. These remarks confirmed the dependence of absorbance at 260 nm for ABMAP as a function of molar ratio nRu(III)/nABMAP (Fig. 8c). In the arrangement studied, the values of molar ratio equal 0.5 which is consistent with stoichiometry 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 of the complex formation.
2 of the complex formation.
The proposed complexing equilibria models correspond to the formation of two Ru(III) ion–ligand complex of stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (metal
2 (metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand) (eqn (1) and (2)). Based on the results from spectrophotometric measurements the values of gradual and cumulative formation constants for Ru(III) complexes were determined (Table 7).
ligand) (eqn (1) and (2)). Based on the results from spectrophotometric measurements the values of gradual and cumulative formation constants for Ru(III) complexes were determined (Table 7).
| Complex cation | Molar ratio | Spectrophotometric technique | |||||
|---|---|---|---|---|---|---|---|
| MeCN | H2O | ||||||
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K1 K1 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K2 K2 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β β |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K1 K1 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K2 K2 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β β |
||
| a Not found or undeterminable. | |||||||
| [Ru(PAOX)2(OH2)2]3+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
4.55 (±0.05) | 4.49 (±0.06) | 9.04 | 3.61 (±0.07) | 7.15 (±0.12) | 10.76 |
| [Ru(DPP)2(OH2)2]3+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
6.80 (±0.02) | 3.97 (±0.02) | 10.77 | 5.17 (±0.16) | 6.46 (±0.19) | 11.63 |
| [Ru(PTCA)2(OH2)2]3+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
4.54 (±0.02) | 4.44 (±0.03) | 8.98 | 4.79 (±0.11) | 8.54 (±0.17) | 13.33 |
| [Ru(ABMAP)2(OH2)2]3+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
4.84 (±0.01) | 4.64 (±0.03) | 9.48 | 4.65 (±0.13) | —a | 4.65 |
Analyzing the values of cumulative stability constants of complexes in acetonitrile, it can be noticed that the most stable complex with ruthenium(III) ion is formed by 2,3-bis(2-pyridyl)pyrazine, while the weakest connection is formed by pyrazine-2-thiocarboxamide. The octahedral coordination is occupied by one water molecule, chlorine atom and azomethine nitrogen and nitrogen of the oxime or amine group in the case of PAOX and PTCA, respectively. The proposed binding set of Ru(III) complex with 2,3-bis-(2-pyridyl)pyrazine includes nitrogen of the azomethine group and pyridyl ring. In case of ABMAP, the central atom is coordinated by two nitrogen atoms of amine and methylamine groups, respectively. During complexation processes more energetically favorable five-membered rings are formed, which is consistent with indicated coordination sites.
The stability constant values of monosubstituted pyrazine derivatives complexes in MeCN (PAOX and PTCA) increase with the number of donor atoms introduced to the substituents in the second position. Furthermore, the same trend can be observed when the increased number of substituents are introduced to the pyrazine ring (ABMAP and DPP). Di-substituted pyrazine derivatives have higher stability constants, which suggests that there occurs the establishment of coordination center consisting of two electron donating groups. Ruthenium(III) ion can bind to chelating elements of pyrazine derivatives, which is demonstrated in the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K1 values. Lower log
K1 values. Lower log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K2 values (lower than log
K2 values (lower than log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K1) resulted from the binding second pyrazine derivative to the Ru(III) ion, decreasing the number of unoccupied places in the coordination sphere of the central atom.
K1) resulted from the binding second pyrazine derivative to the Ru(III) ion, decreasing the number of unoccupied places in the coordination sphere of the central atom.
Our experimental stability studies in aqueous solutions prove that one stability constant can be determined for [Ru(ABMAP)(OH2)4]Cl3 and two for the other complexes. The existence of only one equilibrium in the case of [Ru(ABMAP)(OH2)4]Cl3 is probably caused by both steric and electroinductive effects. Bromide anion is the biggest and most negative substituent in the pyrazine ring among the derivatives studied in this work.
Biological assay – microbiological studies
The presented study was performed to estimate the influence of RuCl3·1.5H2O and pure ligands (PAOX, PTCA, DPP and ABMAP) on growth and multiplication of selected bacterial strains and Candida. MIC and MBC tests were used to compare antibacterial and antifungal activity of Ru(III) complexes and 2 commercial antibiotics – Ciprofloxacin (for bacteria) and Fluconazole (for Candida). The presented results indicated that RuCl3·1.5H2O, pure ligands (PAOX, PTCA, DPP, ABMAP) and Ru(III) complexes in a concentration from 2 to 200 μg mL−1 (Table 8) have no antimicrobial activity.| Compound | Bacteria G (+) | Bacteria G (−) | ||||||
|---|---|---|---|---|---|---|---|---|
| Enterococcus faecalis | Staphylococcus aureus | Klebsiella pneumoniae | Pseudomonas aeruginosa | |||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| μg mL−1 | ||||||||
| RuCl3·1.5H2O | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| PAOX | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| PTCA | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| DPP | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| ABMAP | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| [RuCl(PAOX)2(OH2)]Cl2 | 40 | >200 | 100 | >200 | 100 | >200 | 100 | >200 |
| [RuCl(PTCA)2(OH2)]Cl2 | 100 | >200 | >200 | >200 | >200 | >200 | 150 | >200 |
| [RuCl2(ABMAP)2]Cl | 100 | >200 | >200 | >200 | >200 | >200 | 150 | >200 |
| [RuCl(DPP)(OH2)3]Cl2 | 100 | >200 | >200 | >200 | >200 | >200 | 150 | >200 |
| Ciprofloxacin | 0.5 | 2 | 16 | >256 | <0.06 | <0.06 | 1 | 2 |
In this case, only the [RuCl(PAOX)2(OH2)]Cl2 complex exhibited low activity against tested Gram (−) bacteria in higher concentrations (MIC = 100 μg mL−1) and Gram (+) bacteria: E. faecalis (MIC = 40 μg mL−1) and S. aureus (MIC = 100 μg mL−1). The [RuCl(PAOX)2(OH2)]Cl2 complex tested against C. albicans presented strong antifungal activity (Table 9), i.e. at the level of 5 μg mL−1, which was similar to Fluconazole. Although the MIC of [RuCl(PAOX)2(OH2)]Cl2 for the ATCC 90028 strain was 75 μg mL−1, combining this Ru complex with Fluconazole results in their synergistic antifungal activity. Combination of 4.7 μg mL−1 [RuCl(PAOX)2(OH2)]Cl2 and 0.62 μg mL−1 Fluconazole inhibited the growth of C. albicans (data not shown). Antifungal activity of [RuCl(PAOX)2(OH2)]Cl2 complex may be caused by structural and steric similarity to ‘Ruthenium Red’ impurities like [X(NH3)4Ru(III)ORu(IV) (NH3)4X]3+ (where X = OH−, Cl−) that are known to block Ca2+ uptake in mitochondria blocking the cell respiration process.77 The chelation of the Ru3+ ion by a Schiff base reduces its polarity and enables the complex to pass the phospholipid cell membrane because of the partial shift in Ru charge.78
| Compound | Candida albicans | ||
|---|---|---|---|
| ATCC 14053 | ATCC 90028 | ATCC 10231 | |
| Minimum inhibitory concentration MIC (μg mL−1) | |||
| [RuCl(PAOX)2(OH2)]Cl2 | 10 | 75 | 5 |
| Fluconazole | 5 | 2.5 | 5 |
The salt (RuCl3·1.5H2O), ligands (PAOX, PTCA, DPP, ABMAP) and other Ru(III) complexes presented low antifungal activity (MIC > 200 μg mL−1, data not shown). The yeast C. albicans is by far the most common human pathogenic Candida species and can cause a broad spectrum of diseases including skin, mucosal and systemic infections (candidiasis).79 The antifungal agents available to treat fungal infections are limited. The clinical usefulness of drugs is hampered by their safety (undesirable side effects on patients are often associated with antifungal drugs).80 Therefore, the development of new antifungal drugs is critical to continued effective therapy.
Biological assay – cytotoxicity studies
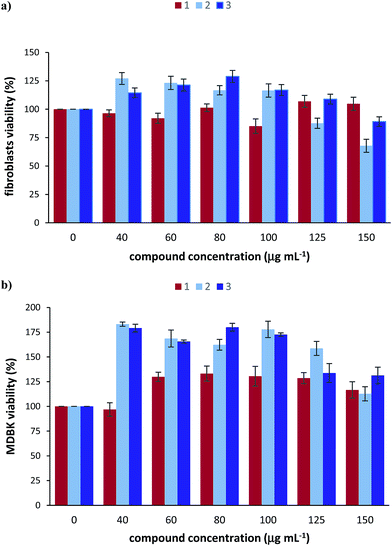
The cytotoxicity of the [RuCl(PAOX)2(OH2)]Cl2 complex as well as RuCl3·1.5H2O and PAOX was investigated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell viability assay that produce coloured formazan in the presence of living cells. The results obtained from this assay are as follows.The cytotoxicity of the three tested compounds in bovine primary fibroblasts was assessed (Fig. 9a). RuCl3·1.5H2O caused about 15% loss of cell viability in concentration of 100 μg mL−1. PAOX seems to stimulate cell proliferation in concentrations range 40–100 μg mL−1 but it appears to have a dose-dependent toxic effect at the higher concentrations (125 μg mL−1 and 150 μg mL−1) with a significant loss of approximately 30% cell viability when the compound was introduced to the cells for 24 hours. [RuCl(PAOX)2(OH2)]Cl2 complex caused slight cytotoxic effect (approximately 10%) only at the highest tested concentration of 150 μg mL−1. Data collected from the cell viability assay of the bovine MDBK cells show that none of the three tested compounds cause a cytotoxic effect, even in very high doses (Fig. 9b). What is more, it is evident that the cell viability in the presence of [RuCl(PAOX)2(OH2)]Cl2 is over 100% after 24 h, indicating that it has proliferative effect on the tested cells.
Biological assay – erythrocyte binding assay
To minimize interference from haemolysis, the spectra were analysed between 240 and 400 nm. For [RuCl(PAOX)2(OH2)]Cl2 the best linearity in the investigated range was found to be at 282 nm (R2 = 0.9965). The concentration of [RuCl(PAOX)2(OH2)]Cl2 in samples was determined with the use of the aforementioned wavelength. The obtained data suggest that the binding of [RuCl(PAOX)2(OH2)]Cl2 to erythrocytes is a concentration dependant in an exponential manner. The obtained binding equation was y = 6.25 × 100.0184x (R2 = 0.9662).Based on the obtained data it can be determined that low concentrations (10 μg mL−1) do show significant binding to erythrocytes. After transmission through the cell membrane, Ru3+ ion could be reduced to Ru2+ ion that could compete for Fe2+ position in HEM. [RuCl(PAOX)2(OH2)]Cl2 complex was deemed effective against fungi in lower ranges than those that show strong binding to erythrocytes.
Conclusions
The choice of the pyrazine derivative ligands for the present investigation was based on the consideration that the Ru(III) ion (relatively hard Lewis acid) had high affinity for hard donor atom (O, N) groups in order to form stable complexes. From the analytical data and the physical studies discussed above, the selected pyrazine derivatives showed bidentate (bis-chelating) coordination mode. The synthesis of Ru(III) with PTCA, PAOX, DPP and ABMAP has already been described in this paper. The binding set of three compounds included azomethine nitrogen and amine nitrogen. The central atom is a coordinated nitrogen atom of the azomethine group and pyridyl ring in [RuCl(DPP)(OH2)3]Cl. The coordination number of the Ru(III) complexes was 6, and on the basis of this coordination number, the structures proposed for the complexes were shown in Fig. 4. The geometries of the obtained complexes were found octahedral, which was consistent with the results of spectroscopic investigations. The 1H NMR and ESI-MS results complemented presented characterization of the Ru(III) complexes as having octahedral species with a selected, bidentate pyrazine derivatives as ligand. The obtained analytical data were consistent with the proposed formula. The results of semi-empirical calculations showed that each Ru(III) complex with selected pyrazine derivative had octahedral geometry, which was in conformity with the data obtained from spectroscopic and analytical experiments. X-ray crystallographic investigations, which might confirm the proposed structures, could not be carried out as we failed to grow suitable crystals of any of these complexes.The present study showed that coordination by the nitrogen donor atoms (amine, amide, and other nitrogen-containing groups) in the bidentate ligands could successfully stabilize the higher oxidation states of a transition metal. This was manifested in the stabilization of trivalent state of ruthenium in the [RuCl(PAOX)2(OH2)]Cl2, [RuCl(DPP)(OH2)3]Cl2, [RuCl(PTCA)2(OH2)]Cl2, and [RuCl2(ABMAP)2]Cl complexes. The electrochemically generated one-electron oxidized and reduced species, viz. [RuIVCl(L)n] and [RuIICl(L)n] (where L denotes PTCA and DPP inside complexes) appeared to have a potential to serve as mild oxidants and reductants, respectively, and such possibilities are currently under study.
The presence of slight bathochromic and hypsochromic shifts combined with hyperchromic effect confirmed the interaction of metal ion with ligand. Spectrophotometric titrations results showed the presence of two Ru(III) ion–ligand complexes of metal–ligand stoichiometry equal 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1
1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. The results obtained from experimental measurements enabled us to conclude that the complexes of ruthenium(III) examined in this work were found stable both in the solid state and solution, which was confirmed by the values of gradual and cumulative stability constants. They can be ordered in accord with their increasing stability:
2. The results obtained from experimental measurements enabled us to conclude that the complexes of ruthenium(III) examined in this work were found stable both in the solid state and solution, which was confirmed by the values of gradual and cumulative stability constants. They can be ordered in accord with their increasing stability:
| [Ru(PTCA)2(OH2)2]3+ > [Ru(PAOX)2(OH2)2]3+ > [Ru(ABMAP)2(OH2)2]3+ > [Ru(DPP)2(OH2)2]3+. |
In this rank Ru(III) forms a complex of the highest stability with 2,3-bis(2-pyridyl)pyrazine, while with pyrazine-2-thiocarboxamide – the weakest one.
Among the coordination compounds of Ru(III) only the complex [RuCl(PAOX)2(OH2)]Cl2 presented strong antifungal activity. The complex causes no adverse health effects and may be used as a potential antifungal agent.
Experiment protocols
Material and methods
All chemicals of analytical purity grade were purchased from Sigma-Aldrich Co. Ltd.: pyrazine-2-amidoxime (pure 97%), pyrazine-2-thiocarboxamide (pure 97%), 2,3-bis-(2-pyridyl)pyrazine (pure 98%), 2-amino-5-bromo-3-(methylamino)pyrazine (pure 97%), ruthenium(III) chloride hydrate (pure 99,98%). All reagents were used without further purification. The solutions were prepared with Hydrolab-Reference-purified water. The percentage compositions of the elements (C, H, N, S) of the synthesized compounds were determined using an element analyzer Carlo Erba EA 1108 CHNS. Infrared spectra of Ru(III) complexes were recorded as potassium bromide (KBr) pellets using Bruker Infrared Spectrometer in the range of 4000–400 cm−1. The 1H NMR spectra of Ru(III) complexes were recorded on a Bruker AVANCE III 500 MHz instrument in DMSO-d6. The chemical shifts values were reported in ppm (δ) and were applied indirectly to tetramethylsilane as a signal of solvent (2.49 for 1H in DMSO-d6). ESI-MS spectra of Ru(III) complexes were recorded in positive ion mode by direct injection at a 5 μL min−1 flow rate using a Bruker Daltonics HCT Ultra high-resolution mass spectrometer equipped with conventional ESI source. All measurements were performed at room temperature.The electronic absorption spectra were recorded in MeCN on a Perkin Elmer Lambda 650 spectrophotometer in the range of 250–700 nm with a spectral band width of 2 nm. The determination of the composition and stabilities of the examined ruthenium(III) complexes were performed with the spectrophotometric titration method in the range of 200–600 nm. The solutions of the studied pyrazine derivatives and metal Ru(III) cation were prepared directly before measurements and were maintained at a constant temperature of 25 °C. The spectrophotometric titrations were carried out at a constant ligand concentration. The concentration of metal ion was about 20 times higher than ligand. Conductivity measurements were obtained using an ELMETRON CC-401 conductivity meter, at 25 °C in water as a solvent, using concentrations of 10−3 mol·dm−3 for all synthesized complexes. Thermal decompositions were measured by means of a thermal equalizer TG209 Netzsch. All experiments were carried out in argon atmosphere. The analyzer was equipped with a programmed temperature controller, which automatically maintains constant temperature during thermal events. The TG weight loss was measured from 20 to 850 °C at a heating rate of 15°C min−1. Infrared spectra were registered in Nujol mulls using a Bruker IFS 66 spectrophotometer. All the measurements were verified at least twice.
Synthesis of the complexes
Ruthenium(III) chloride hydrate (0.41 g; 1.75 mmol), lithium chloride (4.19 g; 100.00 mmol) and 0.49 g (3.52 mmol) pyrazine-2-thiocarboxamide (0.48 g – 3.47 mmol of pyrazine-2-amidoxime) were dissolved in 50 mL dimethylformamide. The mixture was heated at reflux for 8 h and stirred magnetically throughout this period. After the solution was cooled down to room temperature, 250 mL acetone was added and the resulting solution was cooled in a fridge (at 0 °C) overnight. Filtering yielded a red-black solution and a dark brown microcrystalline product for PTCA and a blue-black solution and a dark navy blue microcrystalline product for PAOX. The precipitate was washed three times with 25 mL portions of diethyl ether and then left to slow evaporation at room temperature. In a similar way – with the use of 0.29 g (1.24 mmol) ruthenium(III) chloride hydrate, lithium chloride (4.19 g; 100.00 mmol) and 0.59 g (2.52 mmol) DPP (0.51 g; 2.52 mmol) ABMAP – the black powder of [RuCl(DPP)(OH2)3]Cl2 complex and the black-green powder of [RuCl2(ABMAP)2]Cl were collected after a few days. Their solubilities in water, acetonitrile and DMSO were confirmed for all synthesized complexes.Electrochemical study
All cyclic voltammetry (CV) experiments were performed under inert gas atmosphere. Voltammetric recordings were made using a Gamry Instruments 600 electrochemical analyzer. A platinum-bead working, platinum-coil counter electrode and saturated calomel reference electrode (SCE) were used for CV measurements. Cyclic voltammograms were recorded under argon gas atmosphere in MeCN at room temperature. The concentration of the complexes was 1 mM in the electrolyte solution, where commercially obtained [n-(C4H9)4N]ClO4 (TBAP) was used as a supporting electrolyte. The voltage scan rate during the CV measurements was 100 mV s−1. Controlled potential electrolysis at the anodic peak potential was carried out with an Ag-wire reference electrode after making a correction between the SCE and the Ag-wire (280 mV for SCE).Molecular modeling
Geometry optimization has been performed with the use of semi-empirical PM3 and AM1 methods81 using the Hyperchem 8.82 The correct stereochemistry was assured through the manipulation and modification of the molecular coordinates to obtain reasonable low energy molecular geometries at AM1 and PM3, (Polak-Ribiere) RMS 0.01 kcal.Antimicrobial and antifungal activity
We studied the antimicrobial and antifungal activity of Ru(III) complexes with PAOX, PTCA, DPP, ABMAP and pure ligands (PAOX, PTCA, DPP, ABMAP) and the metal salt RuCl3·1.5H2O within wide concentration ranges (2.5–200 μg mL−1).We used the following strains either resistant or susceptible to antibiotics from 4 species of human bacterial pathogens (hospital isolates obtained from the Laboratory of Microbiology of the Provincial Hospital in Gdansk, Poland are stored in the Faculty of Biotechnology, UG & MUG, Poland): Enterococcus faecalis G (+), Staphylococcus aureus G (+), Klebsiella pneumoniae G (−) and Pseudomonas aeruginosa G (−). The isolates were tested for resistance to 12 antibiotics from the classes of: β-lactams, aminoglycosides, glycopeptides macrolide, fluoroquinolone and lincosamide.83 We also tested 3 strains of Candida albicans (ATCC 14053, ATCC 90028 and ATCC 10231) susceptible to Fluconazole (ThermoFisher, Germany). The bacteria and fungi were grown on BHI medium (BTL, Poland) at 37 °C. The minimum bactericidal concentrations (MBC) of the tested compounds required to achieve the desired antimicrobial effect in planktonic culture were determined using the method described previously.84 For fungi the minimum inhibitory concentration (MIC) was determined using the broth microdilution method according to CLSI method with the inoculum of 2.5 × 103 CFU mL−1. Additional tests were performed for Candida, involving microdilution broth checkerboard techniques in vitro which established pharmacodynamic interactions between [RuCl(PAOX)2(OH2)]Cl2 and the antibiotic Fluconazole, according to the method described by Meletiadis et al.85
Cell culture and treatment with compounds
Primary bovine fibroblasts isolated from calves' ear was cultured in a complete cell culture medium, consisting of DMEM (Dulbecco's Modified Eagle Medium, Sigma) supplemented with 15% heat-inactivated fetal bovine serum (FBS, Gibco), 2 mM L-glutamine (Invitrogen) and 10 mL L−1 of Antibiotic Antimycotic Solution (Sigma), at 37 °C in a 5% CO2 humidified atmosphere. Madin-Darby bovine kidney (MDBK) cells (American Type Culture Collection, ATCC) were maintained at the same conditions in RPMI-1640 medium (Roswell Park Memorial Institute, Sigma) supplemented with 8% heat-inactivated fetal bovine serum, 2 mM L-glutamine and 10 mL L−1 of Antibiotic Antimycotic Solution (Sigma). Stock solutions of investigated compounds (10 mg mL−1) were prepared in dimethyl sulfoxide (DMSO, PanReac AppliChem). Prior to the introduction of tested compounds, 100 μL per well of cells with a cell density of 7.5 × 103 cells per well were seeded in 96-well plates for 24 h. Then the medium was removed and 100 μL of the different concentrations of RuCl3·1.5H2O, PAOX or [RuCl(PAOX)2(OH2)]Cl2 solutions prepared in complete medium from 10 mg mL−1 stocks were added to the cells in each well and incubated for another 24 hours. The control consisted of only the cells and culture medium with the volume of DMSO equal to the volume of suspended compounds in their highest tested concentration.MTT assay
MTT stock solution (5 mg mL−1) was prepared in 1× PBS. Upon exposing the fibroblasts or MDBK cells to the investigated compounds suspensions for 24 h, the MTT solution (20 μL) was added to each well and incubated at 37 °C under 5% CO2 for 3.5 hours. The MTT solution was removed and replaced with MTT solvent (150 μL) to dissolve the insoluble purple formazan crystals produced by the living cells. The plates were covered with tinfoil and gently agitated on an orbital shaker for 15 min, after which they were placed on a plate reader to measure the absorbance at 570 nm. The effects of the tested compounds cell viability were calculated using cells treated with DMSO as control (set as 100%). All experiments were repeated three times.Erythrocyte binding assay
A 2% solution of 100% sheep red blood cells (BioCity Nottingham, United Kingdom) in physiological salt (0.9% NaCl) was freshly prepared before use. The solution was spiked with [RuCl(PAOX)2(OH2)]Cl2 to a final concentration in the range between 5 and 150 μg mL−1. Samples were incubated in 37 °C for 1 hour. After the incubation they were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RCF (relative centrifugal force) for 10 minutes. The supernatant was collected and analyzed using a Perkin Elmer Lambda 25 spectrophotometer in 200–700 nm range. The obtained spectra were used to determine the concentration of unbound [RuCl(PAOX)2(OH2)]Cl2 in the supernatant. Seven point three level calibration curve for [RuCl(PAOX)2(OH2)]Cl2 was performed prior to sample analysis. All experiments were repeated in triplicate.
000 RCF (relative centrifugal force) for 10 minutes. The supernatant was collected and analyzed using a Perkin Elmer Lambda 25 spectrophotometer in 200–700 nm range. The obtained spectra were used to determine the concentration of unbound [RuCl(PAOX)2(OH2)]Cl2 in the supernatant. Seven point three level calibration curve for [RuCl(PAOX)2(OH2)]Cl2 was performed prior to sample analysis. All experiments were repeated in triplicate.
Acknowledgements
The authors gratefully acknowledge the financial support from the Polish Ministry of Science and Higher Education [grant number BMN 538-8236-B698-15], [grant number BMN 538-8236-B663-15], [DS/530-8236-D601-15] and [DS/530-M035-D568-15]. The support of Prof. Zbigniew Kaczyński from the Laboratory of Structural Biochemistry at the Faculty of Chemistry University of Gdańsk in discussion of ESI-MS spectra is greatly appreciated.References
- R. E. Morris, R. E. Aird, P. S. Murdoch, H. Chen, J. Cummings, N. D. Hughes, S. Parson, A. Parkin, G. Boyd, D. I. Jodrell and P. J. Sadler, J. Med. Chem., 2001, 44, 3616 CrossRef CAS PubMed.
- M. Galanski, V. B. Arion, M. A. Jakupec and B. K. Keppler, Curr. Pharm. Des., 2003, 9, 2078 CrossRef CAS PubMed.
- P. Pigeon, S. Top, A. Vessières, M. Huché, E. A. Hillard, E. Salomon and G. Jaouen, J. Med. Chem., 2005, 48, 2814 CrossRef CAS PubMed.
- X. Meng, M. L. Leyva, M. Jenny, I. Gross, S. Benosman, B. Fricker, S. Harlepp, P. Hébraud, A. Boos, P. Wlosik, P. Bischoff, C. Sirlin, M. Pfeffer, J. Loeffler and C. Gaiddon, Cancer Res., 2009, 69, 5458 CrossRef CAS PubMed.
- V. Brabec and O. Nováková, Drug Resist. Updates, 2006, 9(111), 1368 Search PubMed.
- C. S. Allardynce and P. J. Dyson, Platinum Met. Rev., 2001, 45, 62 Search PubMed.
- R. L. Williams, N. H. Toft, B. Winkel and K. J. Brewer, Inorg. Chem., 2003, 42, 4394 CrossRef CAS PubMed.
- A. Bergamo and G. Sava, Dalton Trans., 2007, 13, 1267 RSC.
- C. G. Hartinger, M. A. Jakupec, S. Zorbas-Seifried, M. Groessl, A. Egger, W. Berger, H. Zorbas, P. J. Dyson and B. K. Keppler, Chem. Biodiversity, 2008, 5, 2140 CAS.
- C. G. Hartinger, S. Zorbas-Seifried, M. A. Jakupec, B. Kynast, H. Zorbas and B. K. Keppler, J. Inorg. Chem., 2006, 100, 891 CAS.
- G. Sava, E. Alessio, A. Bergamo and G. Mestroni, Top. Biol. Inorg. Chem., 1999, 1, 143 CAS.
- G. Sava, I. Capozzi, K. Clerici, G. Gagliardi, E. Alessio and G. Mestroni, Clin. Exp. Metastasis, 1998, 16, 371 CrossRef CAS PubMed.
- A. Levina, A. Mitra and P. A. Lay, Metallomics, 2009, 1, 458 RSC.
- L. Trynda-Lemiesz, A. Karaczyn, B. K. Keppler and H. Kozlowski, J. Inorg. Biochem., 2000, 78, 341 CrossRef CAS PubMed.
- O. Mazuryk, K. Kurpiewska, K. Lewinski, G. Stochel and M. Brindell, J. Inorg. Biochem., 2012, 116, 11 CrossRef CAS PubMed.
- A. Calzolari, I. Oliviero, S. Deaglio, G. Mariani, M. Biffoni, N. M. Sposi, F. Malavasi, C. Peschle and U. Testa, Blood Cells, Mol., Dis., 2007, 39, 82 CrossRef CAS PubMed.
- S. Kapitza, M. A. Jakupec, M. Uhl, B. K. Keppler and B. Marian, Cancer Lett., 2005, 226, 115 CrossRef CAS PubMed.
- K. S. Smalley, R. Contractor, N. K. Haass, A. N. Kulp, G. E. Atilla-Gokcumen and D. S. Williams, Cancer Res., 2007, 67, 209 CrossRef CAS PubMed.
- W. H. Ang, A. Casini, G. Sava and P. J. Dyson, J. Org. Chem., 2011, 696, 989 CrossRef CAS.
- M. R. Kamal and R. Levine, J. Org. Chem., 1962, 27, 1360 CrossRef CAS.
- D. Pancechowska-Ksepko, H. Foks, M. Janowiec and Z. Zwolska-Kwiek, Acta Pol. Pharm., 1988, 45, 193 CAS.
- W. Rudnicka, H. Foks, M. Janowiec and Z. Zwolska-Kwiek, Acta Pol. Pharm., 1986, 43, 523 CAS.
- M. H. Cynamon and S. P. Klemens, J. Med. Chem., 1992, 35, 1212 CrossRef CAS PubMed.
- K. Gobis, H. Foks, A. Kędzia, M. Wierzchowska, E. Kwapisz, Z. Zwolska and E. Augustynowicz-Kopeć, Acta Pol. Pharm., 2006, 63, 39 CAS.
- M. Dolezal, P. Cmedlova and L. Palek, Eur. J. Med. Chem., 2008, 43, 1105 CrossRef CAS PubMed.
- T. B. Adams, J. Doull and V. J. Ferron, Food Chem. Toxicol., 2002, 40, 429 CrossRef CAS PubMed.
- D. Frederic, M. Giulio, L. Didier, S. Therese, S. Y. Jacques, R. J. Francois and M. Jacqueline, Eur. J. Med. Chem., 2010, 45, 3564 CrossRef PubMed.
- G. G. Dubinina, M. O. Platonov, S. M. Golovach, P. O. Borysko, A. O. Tolmachov and Y. M. Volovenko, Eur. J. Med. Chem., 2006, 41, 727 CrossRef CAS PubMed.
- M. Shailaja, A. Manjula, S. Venkateshwarlu, B. Vittal Rao and A. Anthony, Eur. J. Med. Chem., 2010, 45, 5208 CrossRef PubMed.
- A. Saxena, J. K. Koacher and J. P. Tandon, J. Antibact. Antifungal Agents, 1981, 9, 435 CAS.
- F. Beckford, J. Thessing, J. Woods, J. Didion, N. Gerasimchuk, A. Gonzalez-Sarrias and N. P. Seeram, Metallomics, 2011, 3, 491 RSC.
- A. M. Pizarro, M. Melchart, A. Habtemariam, L. Salassa, F. P. A. Fabbiani, S. Parsons and P. J. Sadler, Inorg. Chem., 2010, 49, 3310 CrossRef CAS PubMed.
- W. Kandioller, C. G. Hartinger, A. A. Nazarov, C. Bartel, M. Skocic, M. A. Jakupec, V. B. Arion and B. K. Keppler, Chem.–Eur. J., 2009, 15, 12283 CrossRef CAS PubMed.
- M. J. M. Cambell, Coord. Chem. Rev., 1975, 15, 279 CrossRef.
- D. R. Williams, Chem. Rev., 1972, 72, 102 Search PubMed.
- R. K. Agarwal and S. Prasad, Turk. J. Chem., 2004, 28, 691 CAS.
- J. A. Streeky, D. G. Pillsburg and D. H. Busch, Inorg. Chem., 1984, 19, 3148 CrossRef.
- V. Chinnusamy and K. Natarajan, Synth. React. Inorg. Met.-Org. Chem., 1993, 23, 889 CrossRef CAS.
- R. C. Saxena, C. L. Jain, R. Benjamin and S. K. Sangal, J. Indian Chem. Soc., 1986, 63, 435 CAS.
- A. Chylewska, M. Ogryzek, R. Hałasa, A. Dąbrowska, L. Chmurzyński and M. Makowski, J. Coord. Chem., 2014, 67, 2885 CrossRef CAS.
- A. Chylewska, K. Turecka, A. Dąbrowska, W. Werel and L. Chmurzyński, Int. J. Adv. Pharm., Biol. Chem., 2013, 2, 454 CAS.
- A. Chylewska, D. Jacewicz, D. Zarzeczańska and L. Chmurzyński, J. Chem. Thermodyn., 2008, 40, 1290 CrossRef CAS.
- A. Chylewska, M. Ogryzek, L. Chmurzyński and M. Makowski, J. Coord. Chem., 2015, 68, 3761 CrossRef CAS.
- A. Chylewska, A. Sikorski, M. Ogryzek and M. Makowski, J. Mol. Struct., 2016, 1105, 96 CrossRef CAS.
- S. C. Tripathi and S. Paul, Stability constants of Ru(III), Rh(III), Pd(III), Os(VIII), Ir(III) and Pt(IV) chelates with O-coumaric acid, J. Inorg. Nucl. Chem., 1973, 35, 2465–2470 CrossRef CAS.
- D. Chatterjee and A. Mitra, Ruthenium Polyaminocarboxylate Complexes. Prospects for their use as metallopharmaceuticals, Platinum Met. Rev., 2006, 50, 2–12 CrossRef CAS.
- M. Delferro, L. Marchìo, M. Tegoni, S. Tardito, R. Franchi-Gazzola and M. Lanfranchi, Synthesis, structural characterisation and solution chemistry of ruthenium(III) triazole-thiadiazine complexes, Dalton Trans., 2009, 3766–3773 RSC.
- N. Ljubijankić, A. Zahirović, E. Turkušić and E. Kahrović, DNA Binding Properties of Two Ruthenium(III) Complexes Containing Schiff Bases Derived from Salicylaldehyde: Spectroscopic and Electrochemical Evidence of CT DNA Intercalation, Croat. Chem. Acta, 2013, 86, 215–222 CrossRef.
- D. Musumeci, L. Rozza, A. Merlino, L. Paduano, T. Marzo, L. Massai, L. Messoric and D. Montesarchio, Interaction of anticancer Ru(III) complexes with single stranded and duplex DNA model systems, Dalton Trans., 2015, 44, 13914–13925 RSC.
- R. K. Agarwal, Pol. J. Chem., 1991, 65, 1211 CAS.
- R. K. Agarwal and S. Prasad, Turk. J. Chem., 2005, 29, 289 CAS.
- A. Natarajan and S. Pragasam, Asian J. Chem., 1995, 7, 757 CAS.
- A. H. Velders, B. van der Geest, H. Kooijman, A. L. Spek, J. G. Haasnoot and J. Reedijk, Eur. J. Inorg. Chem., 2001, 2, 369 CrossRef.
- K. van der Schilden, A. H. Velders, J. G. Haasnoot and J. Reedijk, J. Inorg. Biochem., 2001, 86, 466 Search PubMed.
- H. Gunther, NMR Spectroscopy, John Wiley & Sons Ltd, 2001 Search PubMed.
- V. K. Sharma, A. Srivastava and S. Srivastava, J. Serb. Chem. Soc., 2006, 71, 917 CrossRef CAS.
- A. Garza-Ortiz, P. U. Maheswari, M. Siegler, A. L. Spek and J. Reedijk, Inorg. Chem., 2008, 47, 6964 CrossRef CAS PubMed.
- M. Delferro, L. Marchìo, M. Tegoni, S. Tardito, R. Franchi-Gazzola and M. Lanfranchi, Dalton Trans., 2009, 19, 3766 RSC.
- P. Dolezel and V. Kuban, Chem. Pap., 2002, 56, 236 CAS.
- D. Musumeci, L. Rozza, A. Merlino, L. Paduano, T. Marzo, L. Massai, L. Messori and D. Montesarchio, Dalton Trans., 2015, 44, 13914 RSC.
- S. A. Hofstadler and K. A. Sannes-Lowery, Nat. Rev. Drug Discovery, 2006, 5, 585 CrossRef CAS PubMed.
- F. Lachaud, A. Quaranta, Y. Pellegrin, P. Dorlet, M. F. Charlot, S. Un, W. Leibl and A. Aukauloo, Angew. Chem., 2005, 44, 1536 CrossRef PubMed.
- G. Venkatachalam, S. Matheswaran and R. Ramesh, Indian J. Chem., 2005, 44, 705 Search PubMed.
- V. K. Sharma, S. Srivastava and A. Srivastava, Bioinorg. Chem. Appl., 2007, 2007, 1 CrossRef PubMed.
- C. K. Jorgensen, Acta Chem. Scand., 1956, 10, 518 CrossRef CAS.
- Y. Tanabe and S. Sugano, J. Phys. Soc. Jpn., 1954, 9, 753 CrossRef CAS.
- J. Wang, Y. Zhao, X. Jin, L. Yang and H. Wang, Spectrochim. Acta, Part A, 2014, 122, 649 CrossRef CAS PubMed.
- R. Abu-Eittah and M. M. Hammed, Bull. Chem. Soc. Jpn., 1984, 57, 844 CrossRef.
- S. Chandra, Nano-Metal Chemistry, 1992, 22, 1565 Search PubMed.
- A. Z. El-Sonbati, A. A. El-Bindary, A. El-Dissouky, T. M. El-Gogary and A. S. Hilali, Spectrochim. Acta, Part A, 2002, 58, 1623 CrossRef CAS.
- E. M. Jouad, A. Riou and M. Allain, Polyhedron, 2001, 20, 67 CrossRef CAS.
- R. Ramesh and S. Maheswaran, J. Inorg. Biochem., 2003, 96, 457 CrossRef CAS PubMed.
- L. Mishra, R. Prajapati and K. K. Pandey, Spectrochim. Acta, Part A, 2008, 70, 79 CrossRef PubMed.
- G. P. Puthilibai, S. Vasudhevan, S. K. Rani and G. Rajagopal, Spectrochim. Acta, Part A, 2009, 72, 796 CrossRef CAS PubMed.
- J. Kostrowicki and A. Liwo, Comput. Chem., 1987, 11, 193 CrossRef.
- D. A. Jose, D. K. Kumar, B. Ganguly and A. Das, Org. Lett., 2004, 6, 3445 CrossRef CAS PubMed.
- W. L. Ying, J. Emerson, M. J. Clarke and D. R. Sanadi, Biochemistry, 1991, 30, 4949 CrossRef CAS PubMed.
- M. J. Clarke, Coord. Chem. Rev., 2003, 236, 209 CrossRef CAS.
- G. Moran, D. Coleman and D. Sullivan, in Candida and Candidiasis, ed. R. A. Calderone and C. J. Clancy, ASM Press, 2nd edn, 2012 Search PubMed.
- D. Gozalbo, P. Roig, E. Villamón and M. L. Gil, Curr. Drug Targets: Infect. Disord., 2004, 4, 117 CrossRef CAS.
- N. L. Allinger, J. Am. Chem. Soc., 1977, 99, 8127 CrossRef CAS.
- HyperChem version 8.0, Hypercube Inc., 2002 Search PubMed.
- K. Bogucka, A. Królicka, W. Kamysz, T. Ossowski, J. Łukasiak and E. Łojkowska, Pol. J. Microbiol., 2004, 53, 41 CAS.
- M. Krychowiak, M. Grinholc, R. Banasiuk, M. Krauze-Baranowska, D. Glod, A. Kawiak and A. Królicka, PLoS One, 2014, 9, e115727 Search PubMed.
- J. Meletiadis, S. Pournaras, E. Roilides and T. J. Walsh, Antimicrob. Agents Chemother., 2010, 54, 602 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary data containing infrared spectra of Ru(III) complexes, spectrophotometric titration curves and Hyperchem data associated with this article can be found. See DOI: 10.1039/c6ra03068h |
| This journal is © The Royal Society of Chemistry 2016 |