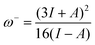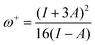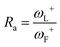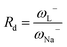Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Theoretical insight into the antioxidant properties of melatonin and derivatives†
Jeffrey R.
Johns
*a and
James A.
Platts
b
aMelatonin Research Group, Faculty of Pharmaceutical Sciences, Khon Kaen University, Khon Kaen, 40002, Thailand. E-mail: jjeff@kku.ac.th; Tel: +66-43-202378
bSchool of Chemistry, Cardiff University, Park Place, Cardiff, CF10 3AT, UK. E-mail: platts@cardiff.ac.uk; Fax: +44 (0)2920-874030; Tel: +44 (0)2920-874950
First published on 19th August 2014
Abstract
Density functional theory calculations on melatonin, metabolites and synthetic derivatives thereof, and a range of other biological antioxidant molecules are presented, with a view to understanding the antioxidant ability of these molecules. After testing of the necessary calculations, we show that melatonin lies close to vitamin E on a donor–acceptor map, indicating that it should be an excellent electron donor but a poor acceptor. The neutral radical metabolite of melatonin is predicted to be an even better donor, whereas other metabolites and synthetic derivatives should retain antioxidant ability but are less powerful than the parent. QSAR models of antioxidant activity, measured in two different assays, are presented. We show that octanol–water partition coefficient is an excellent predictor of activity in lipophilic media, while properties related to electron donor/acceptor power give good fits against activity in aqueous media.
Introduction
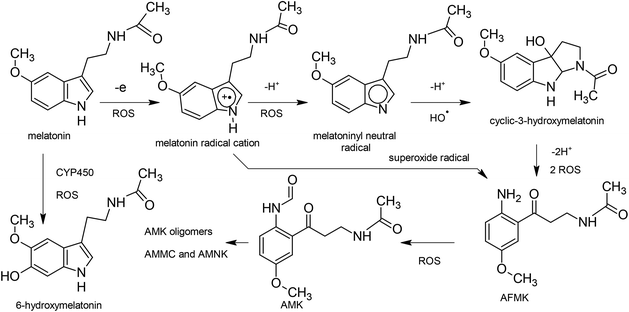
Melatonin (N-acetyl-5-methoxytryptamine) is a naturally occurring molecule, biosynthesized from the precursor amino acid tryptophan, primarily by the pineal gland of vertebrates.1 Melatonin has been extensively reported as a potent antioxidant, both in vitro2 and in vivo.3–5 Much of its effectiveness in vivo may be attributed to the cascade of melatonin antioxidant metabolites produced.6,7 Unlike most small-molecule biological antioxidants such as vitamin C (ascorbic acid), α-tocopherol (vitamin E), lipoic acid etc., melatonin does not redox-cycle. It undergoes molecular rearrangement, effectively removing the free electron from the system – a so-called suicidal antioxidant (Fig. 1). Each of these products of rearrangement is also a potent antioxidant in its own right.2,8,9 Furthermore, most of these processes involve more than one reactive oxygen species (ROS) per step, so that one melatonin molecule could scavenge up to 10 radical species before the final metabolite is eliminated form the body.10 Additionally, the relative position of melatonin and its metabolites in the antioxidant “pecking order” (electrochemical potential) may contribute greatly to its utility in biological systems.11Melatonin is finding great utility in preventing diseases related to oxidative damage including cancer12 and neurodegenerative diseases13,14 as well as its well known role in treatment for reducing insomnia, jet lag, migraine, headache, etc.13,15,16 It is being widely investigated for a large number of other diseases in a large number of clinical trials.17 In addition, consumption of tropical fruits containing melatonin has been shown to reduce antioxidant levels in humans.5,18
The antioxidant radical scavenging properties of melatonin and its metabolites cyclic-3-hydroxymelatonin (cyclic-3OHM),10N(1)-acetyl-N(2)-formyl-5-methoxykynuramine) (AFMK), N(1)-acetyl-5-methoxykynuramine (AMK)7 and 6-hydroxymelatonin (6-OHmel)19 occurs mainly via the one electron transfer process.8,20,21 The first ionization potential (IP) and the electron affinity (EA) are properties of a system that allow measurement of its propensity to donate or accept one electron. The best antioxidants present low IP values, because the lower the IP, the easier the electron abstraction, and vice versa for EA and electron acceptance (antireductant).
Gazquez et al.21 have presented an elegant model to explain relative scavenging activity and antioxidant power of compounds using these two properties. Quantum chemical density functional theory (DFT) calculations can be used to obtain accurate ionization potentials, electron affinities, electrodonating, and electroaccepting power indexes (with respect to internal standards, such as fluorine and sodium atoms). These values can then be used to construct a donator acceptor map (DAM), indicating whether molecules are good electron donors or acceptors. The DAM is a powerful representation of these key properties, helping to reveal the antiradical capacity of any substance and allowing qualitative comparison between substances, alongside quantitative measures obtained from experiment or theory. Previous DAM studies have included linear polyene-conjugated molecules,23 carotenoids,24 a large series of carotenoids,25 carotenoids, melatonin and vitamins,26 and psittacofulvins and anthocyanins.27
Vitamin E or α-tocopherol has been described the “last line of defense” in a multicomponent endogenous antioxidant system.28 It appears that under conditions of stress, depletion of cellular ascorbic acid occurs first, followed by glutathione, then α-tocopherol, resulting in initiation of lipid peroxidation. When glutathione is depleted, ascorbic acid plays a vital role in maintaining cellular α-tocopherol levels and survival of the cell.29 One might expect that melatonin should be depleted after α-tocopherol, particularly in membranes,30 as it is higher in the electrochemical series at 700 mV,8 compared to 500 mV for α-tocopherol.11 Melatonin may therefore truly be the last line of defense against oxidative damage.31 The role of melatonin in this multicomponent antioxidant is still unclear, although there is evidence that melatonin cause upregulation of superoxide dismutase, glutathione reductase and catalase.32,33
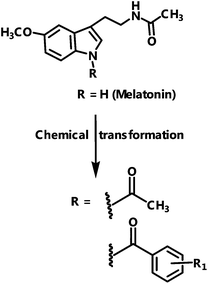
Several melatonin derivatives that were substituted on the indole nitrogen (Fig. 2) have been previously reported for in vitro antioxidant effects and anti-inflammatory activities.34 Their synthesis and characterization is described in this ref. 34.
The aim of this study was to investigate the antioxidant radical scavenging properties of melatonin and its metabolites cyclic-3OHM, AFMK, AMK and 6-hydroxymelatonin, and several N-indole substituted derivatives via the one electron transfer process, using a donor–acceptor map. Other classical antioxidants and vitamins are modeled for comparison. QSAR relationships of some N-indole substituted derivatives between in vitro antioxidant properties experimentally measured by lipid peroxidation of rat brain homogenate using thiobarbituric acid reacting substances (TBARS IC50)35,36 and Oxygen Radical Absorbance Capacity Assay (ORAC) data, and a number of derived electronic properties e.g. HOMO/LUMO energies, donor and acceptor power (Rd, Ra), hardness, electronegativity and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P were investigated. The ORAC assay is based on the scavenging of peroxyl radicals generated by 2,2′-azobis (2-methylpropionamidine) dihydrochloride (AAPH) in aqueous media, which prevent the degradation of the fluorescein probe and, consequently, prevent the loss of fluorescence of the probe.37,38 The antioxidant activity was calculated from the integrated area under the fluorescence curve (AUC) for each antioxidant.
P were investigated. The ORAC assay is based on the scavenging of peroxyl radicals generated by 2,2′-azobis (2-methylpropionamidine) dihydrochloride (AAPH) in aqueous media, which prevent the degradation of the fluorescein probe and, consequently, prevent the loss of fluorescence of the probe.37,38 The antioxidant activity was calculated from the integrated area under the fluorescence curve (AUC) for each antioxidant.
Methods
B3LYP/DFT as implemented in Gaussian09-RevC.0139 software was used for all IP and EA calculations with complete optimizations, without symmetry constraints. Geometries were first minimized in Molecular Operating Environment MOE.40 Calculations were performed on the ARCCA/Raven Supercomputer at Cardiff University. Both vertical IE and EA, where energies for the cation and anion were computed at the optimized geometry of the ground state (single point), and relaxed (adiabatic) IE and EA for optimized cation and anion geometries were calculated. Harmonic frequency analysis was used to verify optimized minima using Molden.41To determine the accuracy of DFT for predicting IP/EA of indoleamines, calculations in gas phase using different basis sets were compared to previously reported photoelectron spectroscopy measurements. IP and bond-dissociation energies for many antioxidant systems do not follow the same trends in gas and solution phases, such that major differences with respect to vacuum are found as when water computations are performed.42 On the basis of the computed BDE and IP values, to more realistically model antioxidant activity in vivo, calculations were therefore performed using the polarizable continuum model (PCM water) i.e. placing the solute in a cavity within the solvent reaction field.
The validity of using B3LYP for calculating EA has been raised, due to most DFT functionals (including B3LYP) being incapable of binding the whole excess electron.43 This may not be revealed when using standard basis sets, even with multiple diffuse functions, since they artificially constrain the electron density to remain near the nuclei. The error due to this constraint depends on the magnitude of the EA, which could render trends in EA unreliable, especially for low EAs. Thus a range-separated DFT method, CAM-B3LYP,44 was used to investigate the problem of fractional EA. A positive energy for the HOMO of an anion species is an easy diagnostic for the fractional EA problem.43
The melatonin molecule contains three freely rotatable bonds in its imidazole side chain. To investigate the effect of conformation, MOE40 was used to select a range of typical conformers via stochastic search of rotatable bonds, using the MMFF94 forcefield. 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 conformers were generated then sorted into clusters based on dihedral angles of the freely rotatable bonds. Vertical IP and EA of the lowest energy conformers from each cluster were then calculated, following geometry optimization of the neutral molecule. No significant changes in geometry on DFT optimization were noted, indicating that conformers remained in their local energy minima.
000 conformers were generated then sorted into clusters based on dihedral angles of the freely rotatable bonds. Vertical IP and EA of the lowest energy conformers from each cluster were then calculated, following geometry optimization of the neutral molecule. No significant changes in geometry on DFT optimization were noted, indicating that conformers remained in their local energy minima.
Donor–acceptor maps were calculated following the method of Martinez et al.,26 using the same experimental values of IE and EA for sodium of 5.140 and 0.540 eV and for fluorine of 17.540 and 3.400 eV respectively, taken from the literature. This set reference points on the map of Ra = 1 for the sodium atom, and Rd = 1 for the fluorine atom. Values calculated using Gaussian 09/RB3LYP/6-31+G* gas phase were 5.406 and 0.596 eV for sodium and 21.405 and 3.513 eV for fluorine respectively.
As formulated by Gazquez et al.22 and applied in the study of Martínez et al.,27 the propensity to donate charge, or electrodonating power, may be defined as:
For electroaccepting power, higher values imply a greater capacity for accepting charge, whereas for electrodonating power, lower values imply a greater capacity for donating charge. It should be noted that ω− and ω+ refer to fractional charges, however I and A refer to donating or accepting a single, whole electron. Thus, a simple charge transfer model, framed in terms of chemical potential and hardness is used to describe electrodonating and electroaccepting powers. The charge flow direction is measured by chemical potential, along with the capacity to donate or accept charge. More emphasis is assigned to the ionization potential than to the electron affinity in the context of the charge donation process. Likewise, more significance is assigned to electron affinity than to ionization potential for electroaccepting power. Hardness provides a measure of the resistance to the electron transfer. So that a range of substances can be compared for electrodonating and electroaccepting power, experimental values of I and A for sodium and fluorine are used a reference points to provide corresponding ω+ and ω− values. Sodium represents a good electron donor and fluorine represents a good electron acceptor. For some substance L, the electron acceptance index can be defined as:
When Ra = 1, then ωL+ ≈ ωF+ and L is as effective an electron acceptor as fluorine. When Ra > 1, then ωL+ > ωF+ and L is a more effective electron acceptor than fluorine. When Ra < 1, then ωL+ < ωF+ and L is a less effective electron acceptor than fluorine. Similarly, the electron donation index can be defined as:
When Rd = 1, then L is as effective an electron donor as sodium, and when Rd > 1, L is a less effective electron donor than sodium, whereas when Rd < 1, L is a more effective electron donor. If Ra and Rd are determined, then any substance L can be characterized in terms of its electron donor–acceptor capacity, and mapped on a donor acceptor map (DAM).
Octanol–water partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) were estimated using ACD/Chemsketch log
P) were estimated using ACD/Chemsketch log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P plugin (Advanced Chemistry Development, Inc., Toronto, Canada, 2012), MarvinView 5.11.3 (Chemaxon, Budapest, Hungary, 2012) and MolKa (Molecular Discovery Ltd, Perugia, Italy, 2012) and compared to literature values where available.
P plugin (Advanced Chemistry Development, Inc., Toronto, Canada, 2012), MarvinView 5.11.3 (Chemaxon, Budapest, Hungary, 2012) and MolKa (Molecular Discovery Ltd, Perugia, Italy, 2012) and compared to literature values where available.
Data on the antioxidant capacity of melatonin derivatives in vitro, using the widely adopted method of Callaway et al.35 for measurement of lipid peroxidation, the thiobarbituric acid reacting substances (TBARS) and brain homogenate, was used in this study. Inhibitory effect on nitric oxide (NO) of melatonin and these derivatives has been previously reported by our group.34
Results and discussion
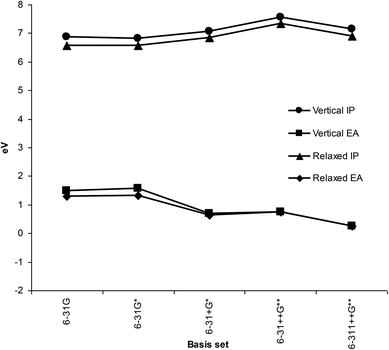
DFT predictions of gas phase IP and EA for different basis sets (Fig. 3) compared favorably with the previously reported photoelectron spectroscopy measurements and IP/EA of other workers (Table 1). Although there were some differences between values obtained with varying basis sets, these were not large. It is notable, though, that smaller basis sets like 6-31G and 6-31G* incorrectly predict positive EA, and that diffuse functions are necessary for qualitatively correct values. Significantly, differences between vertical and relaxed values were small, at any particular basis set. To optimise computational time, the 6-31+G* basis set was chosen for all further calculations as using a larger basis sets did not result in significant improvement in IP or EA values.| 7.76 | IP | PES45 | |
| 7.03 | IP | PES46 | |
| 7.7 | IP | Vertical26 | Estimated from PES |
| 6.83 | IP | Vertical26 | Gaussian03 B3LYP/D95 V gas phase |
| −1.00 | EA | Vertical26 | Gaussian03 B3LYP/D95 V gas phase |
| 7.07 | IP | Vertical | This study. Gaussian09 B3LYP 6-31+G* gas phase |
| −0.70 | EA | Vertical | This study. Gaussian09 B3LYP 6-31+G* gas phase |
| 6.85 | IP | Relaxed | This study. Gaussian09 B3LYP 6-31+G* gas phase |
| −0.65 | EA | Relaxed | This study. Gaussian09 B3LYP 6-31+G* gas phase |
Using the CAM-B3LYP functional did not give significantly different EA values for melatonin using any of the basis sets in this study. Furthermore, no positive energies for HOMO's of any anion species were observed. The reactivity indices calculated for compounds in this study depend mainly on the IP values, rather than EA, as they are mostly electron donors, thus small EA errors will have negligible impact on the overall findings.
Comparison of reported values for IP and EA reported in the literature is made with those calculated using B3LYP 6-31+G* basis set and gas phase in Table 1.
Effect of conformation
Effects of conformation of melatonin on calculated IP and EA in PCM (water) are shown in Table 2. Values of IP and EA calculated in PCM are quite different from those in gas phase due to the effects of solvent polarization. However, conformation changes resulted in less that 1.3% difference in IP and 8.4% difference in EA values in the PCM model. This is not unsurprising, as removal or addition of an electron to the neutral molecule would be expected to affect the extensively delocalized rigid indole moiety only, such that the conformation of the imidazole side chain would have little impact on these processes. Therefore, subsequent DFT calculations reported below use the global energy minimum conformation found from the stochastic search.| MOE conformer energy (kcal mol−1) | Vertical IP/eV | Vertical EA/eV |
|---|---|---|
| 20.573446 | 5.544 | −0.886 |
| 20.573452 | 5.603 | −0.739 |
| 21.250622 | 5.538 | −0.879 |
| 21.250629 | 5.538 | −0.879 |
| 21.694929 | 5.429 | −0.947 |
| 21.694931 | 5.429 | −0.947 |
| 21.724112 | 5.443 | −0.952 |
| Mean | 5.5036 | −0.8898 |
| S.D. | 0.0694 | 0.0745 |
Donor acceptor map
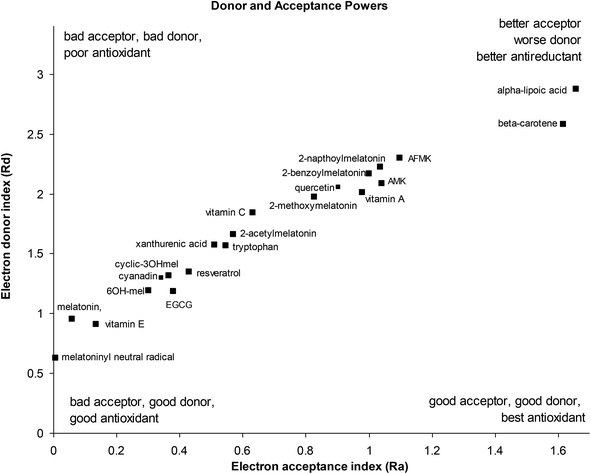
Electron acceptance (Ra) and electron donation (Rd) indexes were calculated from IP and EA, using fluorine and sodium as references22 as previously described in the methods section. The donor acceptor for melatonin, its metabolites and some classical antioxidants is show in Fig. 4.The donor/acceptor maps shows melatonin to be a very good electron donor, along with its metabolites 6-hydroxymelatonin and cyclic-3-hydroxymelatonin and the melatoninyl neutral radical, and several classical antioxidants such as vitamin E (α-tocopherol), epigallocatechin gallate (ECGC), resveratrol, xanthurenic acid and quercetin (a typical flavenoid). Other melatonin metabolites (AFMK, AMK) and melatonin derivatives showed weaker electron donor strength, similar to other classical antioxidants such as vitamin A, vitamin C, beta-carotene and α-lipoic acid. The 4-nitro derivative of melatonin is not shown on the DAM and appears off the top right quadrant at Ra = 4.35 and Rd = 5.62, being a much poorer electron donor and better electron acceptor than melatonin, due to its strongly withdrawing nitro group. Interestingly, the melatoninyl neutral radical that results from a 1-electron 1-proton donation from melatonin is an even more powerful electron donor than melatonin itself.
These results are supported by a large number of experimental observations where melatonin acts as a direct scavenger of free radicals with the ability to detoxify both reactive oxygen and reactive nitrogen species, and indirectly by increasing the activity of the antioxidative defense systems.9,47 Researchers have reported that the peroxyl radical scavenger ability of melatonin is better than of α-tocopherol, vitamin C and reduced glutathione (GSH),48 and more potent than xanthurenic acid, resveratrol, EGCG, vitamin C and α-lipoic acid in inhibiting ˙OH-induced oxidative DNA damage generated by oxygen-derived free radicals from Fenton reaction.49 Melatonin has been demonstrated to reduce the formation of 8-hydroxy-2′-deoxyguanosine, a product of damaged DNA repair, 60 to 70 times more effectively than ascorbate or α-tocopherol.50 Melatonin also plays an important role in protecting cellular membranes against lipid peroxidation.51
It should be noted that most other dietary antioxidants lie outside the scale of this donor–acceptor map, towards the top right quadrant, including carotenoids, psittacofulvins and anthocyanins, flavenoids and polyphenols. These tend to be electron acceptors rather than electron donors i.e. antireductants.
Lipophilicity of antioxidant species
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values for melatonin and its metabolites and some classical antioxidants were calculated and compared to literature values where available (Table 3). The compounds may be classified into roughly three groups – highly lipophilic compounds (log
P values for melatonin and its metabolites and some classical antioxidants were calculated and compared to literature values where available (Table 3). The compounds may be classified into roughly three groups – highly lipophilic compounds (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 6) like α-tocopherol, vitamin A and beta-carotene that mainly protect lipid membranes; vitamin C that is very hydrophilic (log
P > 6) like α-tocopherol, vitamin A and beta-carotene that mainly protect lipid membranes; vitamin C that is very hydrophilic (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < −3) and mainly protects aqueous cellular and tissue environments; and the “melatonin type” compounds that may be considered “amphiphilic” (log
P < −3) and mainly protects aqueous cellular and tissue environments; and the “melatonin type” compounds that may be considered “amphiphilic” (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P between −1 and 2). This latter group should be active antioxidants in all cellular (cytosol and membrane) and tissues environments, and may be important in regenerating some of the other redox-cycling antioxidants like α-tocopherol, and mediating antioxidant reactions at aqueous-lipid membrane interfaces.52 Melatonin has been shown to have strong synergistic effects with α-tocopherol and vitamin C.49,53
P between −1 and 2). This latter group should be active antioxidants in all cellular (cytosol and membrane) and tissues environments, and may be important in regenerating some of the other redox-cycling antioxidants like α-tocopherol, and mediating antioxidant reactions at aqueous-lipid membrane interfaces.52 Melatonin has been shown to have strong synergistic effects with α-tocopherol and vitamin C.49,53
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values
P values
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
||||
|---|---|---|---|---|
| Antioxidant | ACDa | Marvin weightedb | MolKac | Literature values |
a ADC/Chemsketch log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P plugin (Advanced Chemistry Development, Inc., Toronto, Canada) 2012 http://www.acdlabs.com.
b MarvinView 5.11.3 (Chemaxon, Budapest, Hungary) 2012 http://www.chemaxon.com.
c MoKa (Molecular Discovery Ltd, Perugia, Italy) 2012 http://www.moldiscovery.com.
d
http://www.chemicalize.org/structure/#!mol=Vita+E.
e Human Metabolome Database Version 3.6. http://www.hmdb.ca/metabolites/HMDB00305. P plugin (Advanced Chemistry Development, Inc., Toronto, Canada) 2012 http://www.acdlabs.com.
b MarvinView 5.11.3 (Chemaxon, Budapest, Hungary) 2012 http://www.chemaxon.com.
c MoKa (Molecular Discovery Ltd, Perugia, Italy) 2012 http://www.moldiscovery.com.
d
http://www.chemicalize.org/structure/#!mol=Vita+E.
e Human Metabolome Database Version 3.6. http://www.hmdb.ca/metabolites/HMDB00305.
|
||||
| Beta-carotene | 15.51 ± 0.43 | 11.12 | 9.0 | 14.76 |
| α-Tocopherol (vitamin E) | 10.66 ± 0.28 | 8.94 | 9.0 | 10.51d |
| Vitamin A (retinol) | 6.84 ± 0.33 | 6.07 | 6.1 | 4.69–6.38e |
| Alpha-lipoic acid | 2.16 ± 0.29 | 2.11 | 2.4 | — |
| Melatonin | 0.96 ± 0.44 | 1.41 | 1.4 | 1.254,55 |
| 6-OH-melatonin | 0.02 ± 0.83 | 0.84 | 1.0 | — |
| Melatoninyl neutral radical | 0.02 ± 0.83 | 0.88 | — | — |
| AFMK | 0.82 ± 0.52 | 0.34 | 0.0 | 0.4856 |
| AMK | 0.65 ± 0.49 | 0.33 | 0.2 | 0.7456 |
| −0.7758 | ||||
| Tryptophan | 0.87 ± 0.31 | 1.51 | 0.6 | 1.0857 |
| −1.0659 | ||||
| 2-Naphthoyl-melatonin | 2.73 ± 0.46 | 3.31 | 3.7 | — |
| 2-Benzoyl-melatonin | 1.5 ± 0.46 | 2.32 | 2.4 | — |
| Cyclic-3OHmel | −0.64 ± 0.89 | −0.21 | 0.8 | — |
| Acetyl-melatonin | 1.00 ± 0.87 | 0.47 | 1.3 | — |
| Vitamin C | −3.26 ± 0.56 | −1.98 | −3.4 | −1.8558 |
TBARS pIC50 results were correlated with a number of derived electronic properties e.g. HOMO/LUMO energies, donor and acceptor power (Rd, Ra), hardness, electronegativity and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, and results are shown in Table 4. Only lipophilicity (log
P, and results are shown in Table 4. Only lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) correlated well, correlation with other molecular properties (donor/acceptor power, electronegativity and LUMO was poorer, with low or no correlation with hardness, HOMO and HOMO/LUMO energy difference. This may be because of the nature of the brain homogenate lipid peroxidation assay, where solubility of the antioxidant in the lipid domain is the dominant factor contributing to radical scavenging. The hydrophobicity of the antioxidant may also be an important criterion for passive transport into cells across the hydrophobic phospholipid bilayer of the cellular membranes. Furthermore, the single electron transfer mechanism for direct radical scavenging of melatonin, although the most favourable mechanism in aqueous solution, is not favourable in aprotic solvents e.g. benzene, where hydrogen atom transfer/proton coupled electron transfer or radical adduct formation are favoured.29
P) correlated well, correlation with other molecular properties (donor/acceptor power, electronegativity and LUMO was poorer, with low or no correlation with hardness, HOMO and HOMO/LUMO energy difference. This may be because of the nature of the brain homogenate lipid peroxidation assay, where solubility of the antioxidant in the lipid domain is the dominant factor contributing to radical scavenging. The hydrophobicity of the antioxidant may also be an important criterion for passive transport into cells across the hydrophobic phospholipid bilayer of the cellular membranes. Furthermore, the single electron transfer mechanism for direct radical scavenging of melatonin, although the most favourable mechanism in aqueous solution, is not favourable in aprotic solvents e.g. benzene, where hydrogen atom transfer/proton coupled electron transfer or radical adduct formation are favoured.29
| Molecular parameters | Correlation (r2) with TBARS pIC50 | Correlation (r2) with ORAC log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AUC AUC |
|---|---|---|
| Donor power (Rd) | 0.636 | 0.951 |
| Acceptor power (Ra) | 0.647 | 0.935 |
| Hardness (η) | 0.458 | 0.982 |
| Electronegativity (X) | 0.589 | 0.987 |
| Energy HOMO (eV) | 0.331 | 0.928 |
| Energy LUMO (eV) | 0.560 | 0.995 |
| Energy HOMO–LUMO (eV) | 0.237 | 0.941 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
0.978 | 0.554 |
By contrast, for the ORAC assay, which was performed in aqueous medium, all molecular parameters correlated highly with the ORAC AUC except for log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (Table 4), as has been observed previously with indoleamines where antioxidant potency was measured for lipid peroxidation using a conjugated dienes assay60 and in phenolic compounds.61
P (Table 4), as has been observed previously with indoleamines where antioxidant potency was measured for lipid peroxidation using a conjugated dienes assay60 and in phenolic compounds.61
Some QSAR correlation plots are shown in Fig. 5, and all correlation plots are shown in the ESI.†
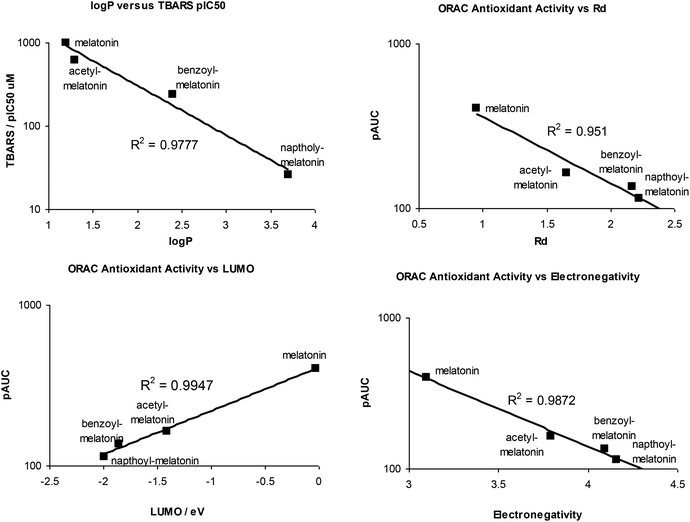 | ||
| Fig. 5 QSAR correlation plots of some calculated molecular parameters and TBARS or ORAC antioxidant activity. | ||
Conclusions
The electron donor power of melatonin and its metabolites demonstrated in this theoretical study support the experimental evidence that melatonin is a powerful biological antioxidant and radical scavenger. Our computational studies, presented above, shed light on this important biological property. We have shown that the B3LYP DFT method, along with the 6-31+G(d) basis set, satisfactorily reproduces experimental gas phase ionization potential and electron affinity, while larger basis sets do not improve performance. Importantly, calculated properties are not dependent on molecular conformation, such that data derived from a single conformation should be sufficient to capture all relevant aspects of this molecule.This method has therefore been used to map out the donor–acceptor power of melatonin, its metabolites, some synthetic derivatives and a range of classical antioxidants. This approach clearly shows that melatonin lies in the range of good electron donors and bad electron acceptors, with similar power to vitamin E. Interestingly, the first neutral, radical metabolite of melatonin is an even better donor than the parent molecule, which will have important implications for the overall biology of the cascade process by which melatonin mops up ROS. Other metabolites, as well as most synthetic derivatives, remain in the range where substantial antioxidant ability should be expected, but a 4-nitro derivative lies well outside this region.
QSAR investigation indicates the ability of melatonin derivatives to protect against lipid peroxidation of brain homogenate strongly correlated with their lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) but only weakly to other molecular properties related to donor/acceptor ability (donor/acceptor power, electronegativity, hardness, HOMO, LUMO, HOMO-LUMO energies). By contrast, these molecular parameters correlate strongly with ORAC antioxidant power measured in aqueous phase.
P) but only weakly to other molecular properties related to donor/acceptor ability (donor/acceptor power, electronegativity, hardness, HOMO, LUMO, HOMO-LUMO energies). By contrast, these molecular parameters correlate strongly with ORAC antioxidant power measured in aqueous phase.
The range of lipophilicity of melatonin and its metabolites (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P between −1 and 2) may explain the large number of antioxidant arenas where melatonin seems to play a role in protecting against ROS damage; they lie between the traditional membrane protectors (α-tocopherol, vitamin A and carotenoids) and hydrophilic compounds (vitamin C, lipoic-acid) and aqueous antioxidant enzymes.
P between −1 and 2) may explain the large number of antioxidant arenas where melatonin seems to play a role in protecting against ROS damage; they lie between the traditional membrane protectors (α-tocopherol, vitamin A and carotenoids) and hydrophilic compounds (vitamin C, lipoic-acid) and aqueous antioxidant enzymes.
Acknowledgements
This work was performed using the computational facilities of the Advanced Research Computing @ Cardiff (ARCCA) Division, Cardiff University and supported by the Melatonin Research Group, Khon Kaen University. Acknowledgement is given to Dr Ploentip Puthongking for synthesis and provision of melatonin derivatives, and Preeaporn Plaimee and Chawapon Pipatwatcharadate for antioxidant measurements.References
- J. H. Stehle, A. Saade, O. Rawashdeh, K. Ackermann, A. Jilg, T. Sebestény and E. Maronde, J. Pineal Res., 2011, 51, 17–43 CrossRef CAS PubMed.
- D. X. Tan, R. Hardeland, L. C. Manchester, B. Poeggeler, S. Lopez-Burillo, J. C. Mayo, R. M. Sainz and R. J. Reiter, J. Pineal Res., 2003, 34(4), 249–259 CrossRef CAS.
- R. J. Reiter, L. C. Manchester and D. X. Tan, Nutrition, 2005, 21(9), 920–924 CrossRef CAS PubMed.
- A. Piechota, S. Lipińska, J. Szemraj and A. Goraca, Gen. Physiol. Biophys., 2010, 29(2), 144–150 CrossRef CAS.
- M. Sae-Teaw, J. Johns, N. P. Johns and S. Subongkot, J. Pineal Res., 2013, 55(1), 58–64 CrossRef CAS PubMed.
- G. R. Martinez, E. A. Almeida, C. F. Klitzke, J. Onuki, F. M. Prado, M. H. Medeiros and P. Di Mascio, Endocrine, 2005, 27(2), 111–118 CrossRef CAS PubMed.
- R. Hardeland, D. X. Tan and R. J. Reiter, J. Pineal Res., 2009, 47(2), 109–126 CrossRef CAS PubMed.
- A. Galano, D. X. Tan and R. J. Reiter, J. Pineal Res., 2013, 54(3), 245–257 CrossRef CAS PubMed.
- D. X. Tan, R. J. Reiter, L. C. Manchester, M. T. Yan, M. El-Sawi, R. M. Sainz, J. C. Mayo, R. Kohen, M. Allegra and R. Hardeland, Curr. Top. Med. Chem., 2002, 2(2), 181–197 CrossRef CAS.
- D. X. Tan, L. C. Manchester, M. P. Terron, L. J. Flores and R. J. Reiter, J. Pineal Res., 2007, 42(1), 28–42 CrossRef CAS PubMed.
- G. R. Buettner, Arch. Biochem. Biophys., 1993, 300(2), 535–543 CrossRef CAS PubMed.
- V. Motilva, S. García-Mauriño, E. Talero and M. Illanes, J. Pineal Res., 2011, 51, 44–60 CrossRef CAS PubMed.
- D. P. Cardinali, V. Srinivasan, A. Brzezinski and G. M. Brown, J. Pineal Res., 2012, 52, 365–375 CrossRef CAS PubMed.
- S. A. Rosales-Corral, D. Acuna-Castroviejo, A. Coto-Montes, J. A. Boga, L. C. Manchester, L. Fuentes-Broto, A. Korkmaz, S. Ma, D. X. Tan and R. J. Reiter, J. Pineal Res., 2012, 52, 67–202 CrossRef PubMed.
- A. Brzezinski, Melatonin in humans, N. Engl. J. Med., 1997, 336, 186–195 CrossRef CAS PubMed.
- R. Hardeland, J. A. Madrid, D. X. Tan and R. J. Reiter, J. Pineal Res., 2012, 52, 139–166 CrossRef CAS PubMed.
- A. Korkmaz, R. J. Reiter, T. Topal, L. C. Manchester, S. Oter and D. X. Tan, Mol. Med., 2009, 15(1–2), 43–50 CAS.
- N. P. Johns, J. Johns, S. Porasuphatana, P. Plaimee and M. Sae-Teaw, J. Agric. Food Chem., 2013, 61(4), 913–919 CrossRef CAS PubMed.
- X. Ma, J. R. Idle, K. W. Krausz and F. J. Gonzalez, Drug Metab. Dispos., 2005, 33, 489–494 CrossRef CAS PubMed.
- A. Galano, Phys. Chem. Chem. Phys., 2011, 13, 7147–7157 Search PubMed.
- A. Galano, D. X. Tan and R. J. Reiter, RSC Adv., 2014, 4, 5220–5227 RSC.
- J. L. Gazquez, A. Cedillo and A. Vela, J. Phys. Chem. A, 2007, 111, 1966–1970 CrossRef CAS PubMed.
- A. Martínez, J. Phys. Chem. B, 2009, 113(10), 3212–3217 CrossRef PubMed.
- A. Martínez, R. Vargas and A. Galano, J. Phys. Chem. B, 2009, 113(35), 12113–12120 CrossRef PubMed.
- A. Galano, J. Phys. Chem. B, 2007, 111(44), 12898–12908 CrossRef CAS PubMed.
- A. Martínez, M. A. Rodríguez-Gironés, A. Barbosa and M. Costas, J. Phys. Chem. A, 2008, 112(38), 9037–9042 CrossRef PubMed.
- A. Martínez, J. Phys. Chem. B, 2009, 113(14), 4915–4921 CrossRef PubMed.
- J. M. May, Z. C. Qu and S. Mendiratta, Arch. Biochem. Biophys., 1998, 349(2), 281–289 CrossRef CAS PubMed.
- S. J. Padayatty, A. Katz, Y. Wang, P. Eck, O. Kwon, J. H. Lee, S. Chen, C. Corpe, A. Dutta, S. K. Dutta and M. Levine, J. Am. Coll. Nutr., 2003, 22, 18–35 CrossRef CAS.
- E. J. Costa, C. S. Shida, M. H. Biaggi, A. S. Ito and M. T. Lamy-Freund, FEBS Lett., 1997, 416(1), 103–106 CrossRef CAS.
- A. Teixeira, M. P. Morfim, C. de Cordova, C. Charão, V. R. de Lima and T. B. Creczynski-Pasa, J. Pineal Res., 2003, 35(4), 262–268 CrossRef CAS.
- C. Rodriguez, J. C. Mayo, R. M. Sainz, I. Antolín, F. Herrera, V. Martín and R. J. Reiter, J. Pineal Res., 2004, 36(1), 1–9 CrossRef CAS.
- J. C. Mayo, R. M. Sainz, I. Antoli, F. Herrera, V. Martin and C. Rodriguez, Cell. Mol. Life Sci., 2002, 59, 1706–1713 CrossRef CAS.
- C. Phiphatwatcharaded, A. Topark-Ngarm, P. Puthongking and P. Mahakunakorn, Drug Dev. Res., 2014, 75(4), 235–245 CrossRef CAS PubMed.
- J. K. Callaway, P. M. Beart and B. Jarrott, J. Pharmacol. Toxicol. Methods, 1998, 39(3), 155–162 CrossRef CAS.
- L. T. Rael, G. W. Thomas, M. L. Craun, C. G. Curtis, R. Bar-Or and D. Bar-Or, J. Biochem. Mol. Biol., 2004, 37(6), 749–752 CrossRef CAS.
- D. Huang, B. Ou and R. L. Prior, J. Agric. Food Chem., 2005, 53, 1841–1856 CrossRef CAS PubMed.
- R. L. Prior, X. Wu and K. Schaich, J. Agric. Food Chem., 2005, 53, 4290–4302 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, N. J. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09 (Revision D.01), Gaussian, Inc., Wallingford, CT, 2009 Search PubMed.
- Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2013.
- G. Schaftenaar and J. H. Noordik, J. Comput. Aided Mol. Des., 2000, 14, 123–134 CrossRef CAS.
- M. Leopoldini, T. Marino, N. Russo and M. Toscano, J. Phys. Chem. A, 2004, 108, 4916–4922 CrossRef CAS.
- F. Jensen, J. Chem. Theory Comput., 2010, 6(9), 2726–2735 CrossRef CAS.
- T. Yanai, D. P. Tew and N. C. Handy, Chem. Phys. Lett., 2004, 393(1–3), 51–57 CrossRef CAS PubMed.
- M. Kubota and T. Kobayashi, J. Electron Spectrosc. Relat. Phenom., 2003, 128(2), 165–178 CrossRef CAS.
- P. H. Cannington and N. S. Ham, J. Electron Spectrosc. Relat. Phenom., 1983, 32, 139 CrossRef CAS.
- R. J. Reiter, D. X. Tan, L. C. Manchester and W. Qi, Cell Biochem. Biophys., 2001, 34, 237–256 CrossRef CAS.
- C. Pieri, M. Marra, F. Moroni, R. Recchioni and F. Marcheselli, Life Sci., 1994, 55(15), PL271–PL276 CrossRef CAS.
- S. Lopez-Burillo, D. X. Tan, J. C. Mayo, R. M. Sainz, L. C. Manchester and R. J. Reiter, J. Pineal Res., 2003, 34, 269–277 CrossRef CAS.
- W. Qi, R. J. Reiter, D. X. Tan, L. C. Manchester, A. W. Siu and J. J. Garcia, J. Pineal Res., 2001, 29, 54–61 Search PubMed.
- A. Catalá, Curr. Mol. Med., 2007, 7(7), 638–649 CrossRef.
- H. Sun, D. V. Greathouse, O. S. Andersen and R. E. Koeppe 2nd, J. Biol. Chem., 2008, 283(32), 22233–22243 CrossRef CAS PubMed.
- E. Gitto, D. X. Tan, R. J. Reiter, M. Karbownik, L. C. Manchester, S. Cuzzocrea, F. Fulia and I. Barberi, J. Pharm. Pharmacol., 2001, 53, 1393–1401 CrossRef CAS.
- L. Kikwai, N. Kanikkannan, R. J. Babu and M. Singh, J. Controlled Release, 2002, 83(2), 307–311 CrossRef CAS.
- M. Mor, C. Silva, F. Vacondio, P. V. Plazzi, S. Bertoni, G. Spadoni, G. Diamantini, A. Bedini, G. Tarzia, M. Zusso, D. Franceschini and P. Giusti, J. Pineal Res., 2004, 36(2), 95–102 CrossRef CAS.
- C. Harthe, D. Claudy, H. Dechaud, B. Vivien-Roels, P. Pevet and B. Claustrat, Life Sci., 2003, 73(12), 1587–1597 CrossRef CAS.
- Liquid Chromatography in Biomedical Analysis, J. Chromatography Library, ed. T. Hanai, Elsevier, New York, 1991, vol. 50 Search PubMed.
- A. Avdeef and K. J. Box, Sirius Technical Application Notes (STAN), Sirius Analytical Instruments Ltd, Forest Row, UK, 1995, vol. 2 Search PubMed.
- M. Urakami, R. Ano, Y. Kimura, M. Shima, R. Matsuno, T. Ueno and M. Z. Akamatsu, Z. Naturforsch C: Biosci., 2003, 58(1–2), 135–142 CAS.
- G. Spadoni, G. Diamantini, A. Bedini, G. Tarzia, F. Vacondio, C. Silva, M. Rivara, M. Mor, P. V. Plazzi, M. Zusso, D. Franceschini and P. Giusti, J. Pineal Res., 2006, 40(3), 259–269 CrossRef CAS PubMed.
- L. V. B. Hoelz, B. A. C. Horta, J. Q. Araújo, M. G. Albuquerque, R. B. de Alencastro and J. F. M. da Silva, J. Chem. Pharm. Res., 2010, 2(5), 291–306 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ob01396d |
| This journal is © The Royal Society of Chemistry 2014 |