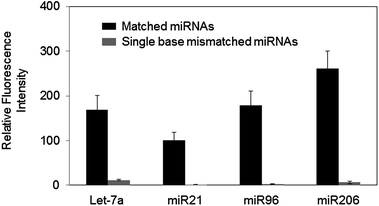
Highly improved specificity for hybridization-based microRNA detection by controlled surface dissociation†
Hye Ryeon
Yoon
ab,
Jeong Min
Lee
c,
Juyeon
Jung
ab,
Chang-Soo
Lee
ab,
Bong Hyun
Chung
*ab and
Yongwon
Jung
*c
aBioNanotechnology Research Center, Korea Research Institute of Bioscience and Biotechnology, P.O. Box 115, Daejeon 305-600, Korea. E-mail: chungbh@kribb.re.kr
bNanobiotechnology, School of Engineering, University of Science and Technology (UST), P.O. Box 115, Daejeon 305-333, Korea
cDepartment of Chemistry, Korea Advanced Institute of Science and Technology, Daejeon 305-701, Korea. E-mail: ywjung@kaist.ac.kr
First published on 18th October 2013
Abstract
Poor specificity has been a lingering problem in many microRNA profiling methods, particularly surface hybridization-based methods such as microarrays. Here, we carefully investigated surface hybridization and dissociation processes of a number of sequentially similar microRNAs against nucleic acid capture probes. Single-base mismatched microRNAs were similarly hybridized to a complementary DNA capture probe and thereby poorly discriminated during conventional stringent hybridization. Interestingly, however, mismatched microRNAs showed significantly faster dissociation from the probe than the perfectly matched microRNA. Systematic analysis of various washing conditions clearly demonstrated that extremely high specificity can be obtained by releasing non-specific microRNAs from assay surfaces during a stringent and controlled dissociation step. For instance, compared with stringent hybridization, surface dissociation control provided up to 6-fold better specificity for Let-7a detection than for other Let-7 family microRNAs. In addition, a synthetically introduced single-base mismatch on miR206 was almost completely discriminated by optimized surface dissociation of captured microRNAs, while this mismatch was barely distinguished from target miR206 during stringent hybridization. Furthermore, a single dissociation condition was successfully used to simultaneously measure four different microRNAs with extremely high specificity using melting temperature-equalized capture probes. The present study on selective dissociation of surface bound microRNAs can be easily applied to various hybridization based detection methods for improved specificity.
Introduction
MicroRNAs (miRNAs) are a class of small noncoding RNAs (19–24 nucleotides, nt) that regulate a considerable portion of gene expression in various species including humans.1 Close to a thousand miRNAs have been identified in humans, and a rapidly expanding body of evidence indicates that miRNAs are promising markers for a number of diseases.2–4 Reliable detection technologies of multiple miRNAs are in great need for fundamental miRNA research as well as medical applications.5 Beside several main commercial techniques for miRNA profiling, such as microarrays, sequencing, and real-time RT-PCR, various novel detection strategies and devices have been developed and applied in miRNA exploration.6–9Many techniques for miRNA profiling rely on surface hybridization between target miRNAs and complementary nucleic acid probes. Mature miRNAs are, however, short and display a wide range of melting temperatures (Tm) against their capture probes. In addition, miRNA family members are often sequentially similar and sometimes differ by only 1 nt in sequence. Most hybridization-based miRNA profiling techniques are challenged by these features of miRNA and often suffer from poor specificity, thus resulting in false signals.5 Low specificity is one of the major sources of inconsistent miRNA profiling results between different detection methods and also between independent measurements, an issue that has been raised in many recent reports.10–12
Multiple strategies have been reported for specific detection of short but sequentially similar miRNAs by careful design of complementary capture probes and optimization of hybridization conditions.6,13,14 In general, capture probes are designed to have equalized Tm values, and hybridization stringency is optimized for adequately high specificity while maintaining sufficient sensitivity. It is generally accepted, however, that similar sequences such as single base mismatches between miRNAs cannot be fully discriminated by conventional surface hybridization.13 Offering a generally applicable solution for enhanced specificity during miRNA surface hybridization is one of the major issues in the field of hybridization-based miRNA profiling.
Previously, we reported an unconventional profiling method for intact miRNAs with two short locked nucleic acid (LNA)-modified probes (∼10 nt).15 miRNAs were first captured on surface bound LNA probes and subsequently detected by a second (highly stringent) hybridization step with the next LNA probes. Interestingly, non-specific miRNAs were highly unstable during this second harsh hybridization with two sandwiching LNA probes, therefore offering unusually high specificity.15 However, a detailed mechanism of this miRNA discrimination during the second hybridization process is still not clear. More importantly, a critical question is whether this two-step/multi-probe hybridization is applicable to general miRNA surface hybridization with a single capture probe for improved specificity.
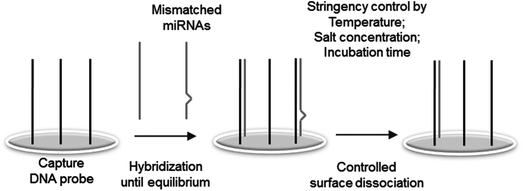
Herein, we demonstrated that significantly improved specificity between target miRNAs and DNA capture probes can be obtained by selective dissociations of bound miRNAs from the assay surface. Sequentially similar (particularly single-base mismatched) miRNAs showed similar bindings to a capture probe during conventional stringent hybridization, resulting in poor specificity. Interestingly, however, these similar miRNAs displayed vastly different dissociation characteristics from the capture probe. Multiple dissociation conditions were determined for highly effective discrimination of many similar miRNAs. Following hybridization between surface bound probes and miRNAs (including mismatched miRNAs) under a low stringent condition, high stringent dissociation conditions (high temperature and/or low salt concentration) were determined to predominantly remove mismatched miRNAs (Fig. 1). Many single-base mismatched miRNAs were effectively discriminated by this controlled dissociation.
Results and discussions
Surface hybridization and dissociation during array detection of Let-7 family miRNAs
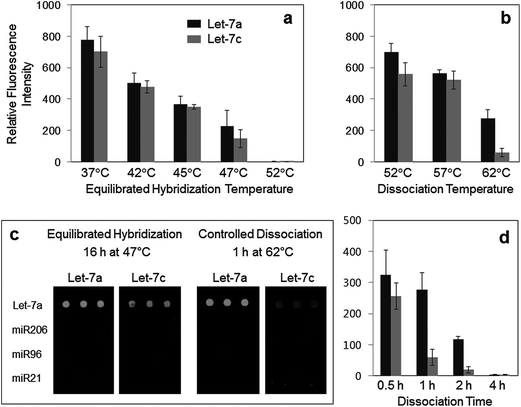
To develop generally applicable hybridization strategies for specific miRNA detection, DNA capture probes were simply designed to recognize entire sequences of target miRNAs (Let-7a, miR21, miR96, and miR206) without any modifications (Table 1). Target miRNAs were synthesized with a biotin moiety, and hybridized miRNAs on surface-spotted DNA capture probes were identified by fluorescence dye-labeled streptavidin. We first investigated hybridization-based detection of human Let-7a amongst highly similar Let-7 family miRNAs (Fig. 2). In particular, Let-7a and singly mismatched Let-7c are among the most difficult miRNA pairs to discriminate.13,16 Biotinylated Let-7a and Let-7c miRNAs (100 pM in 0.1 mL of solution) were applied to a surface-immobilized Let-7a-specific DNA capture probe under various temperatures, ranging from 37 °C to 52 °C. Incubation was continued for 16 h, such that miRNAs were hybridized and simultaneously distinguished until these interactions reached near equilibrium (between association and dissociation). Hybridized glass slides were briefly rinsed with a washing buffer at the temperature that was used during equilibrated hybridization. Specific fluorescence signals of both Let-7a and Let-7c were decreased as hybridization temperatures increased. Even at high temperature, however, Let-7a and Let-7c were not effectively discriminated during this equilibrated hybridization (Fig. 2a). Noticeable discrimination was observed only at 47 °C, and the cross-hybridization of Let-7c was nearly 65%. The binding energies of Let-7a and Let-7c to the DNA capture probe might be too similar, and therefore it is extremely difficult to determine a single equilibrated binding condition (temperature) for effective discrimination of these single-base mismatched miRNAs.| miRNA | miRNA sequence (5′ → 3′) | Capture probe sequence (5′ → 3′) |
|---|---|---|
| a The 5′ ends of all miRNAs contained biotin modifications. Base mismatches are indicated (bold, underlined). The 5′ ends of all capture probes contained amine modifications (amino) and (C)10 linkers. LNA modifications are indicated (italics, underlined). | ||
| miR21 | UAGCUUAUCAGACUGAUGUUGA | DNA: TCAACATCAGTCTGATAAGCTA |
| miR21-mis | UAGCUUAUCAGACUGA![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GUUGA GUUGA |
LNA: T![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) AAC AAC![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) T T![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) A A![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) TCTG TCTG![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) T T |
| miR96 | UUUGGCACUAGCACAUUUUUGCU | DNA: AGCAAAAATGTGCTAGTGCCAAA |
| miR96-mis | UUUGGCACUAGCACAUU![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) UUGCU UUGCU |
LNA: A![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) C C![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) A A![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) A A![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) T T![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) T T![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) CT CT![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) G G |
| miR206 | UGGAAUGUAAGGAAGUGUGUGG | DNA: CCACACACTTCCTTACATTCCA |
| miR206-mis | UGGAAUGUAAGGAAGU![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) UGUGG UGUGG |
LNA: C![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) A A![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) A A![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) A A![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) TTCCTT TTCCTT![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) C C |
| Let-7a | UGAGGUAGUAGGUUGUAUAGUU | DNA: AACTATACAACCTACTACCTCA, LNA: A![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) C C![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) A A![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) CAA CAA![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) CT CT![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) CT CT |
| Let-7b | UGAGGUAGUAGGUUGU![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) U U![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) GUU GUU |
|
| Let-7c | UGAGGUAGUAGGUUGUAU![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) GUU GUU |
|
| Let-7d |
![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) GAGGUAGUAGGUUG GAGGUAGUAGGUUG![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) AUAGU– AUAGU– |
|
| Let-7f | UGAGGUAGUAG![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) UUGUAUAGUU UUGUAUAGUU |
|
We next investigated various dissociation processes of already hybridized Let-7 miRNAs from the Let-7a capture probe. Since the maximum fluorescence signal for Let-7a was detected at 37 °C, miRNAs were first captured on the complementary DNA probe at 37 °C, and then the bound miRNAs (both Let-7a and Let-7c) were washed under highly stringent conditions (52 °C to 62 °C) for 1 h (in 10 mL of solution). A large volume of a washing solution was used to sufficiently reduce the level of re-hybridization of dissociated miRNAs. Interestingly, dissociation at 62 °C offers highly improved specificity for Let-7a detection (Fig. 2b and c). Only ∼21% cross-hybridization of Let-7c was observed, more than 3-fold lower than that of equilibrated hybridization at 47 °C. Dissociation duration at 62 °C was also varied to follow time-dependent dissociation of both Let-7a and Let-7c (Fig. 2d). Gradual increases in the washing time clearly resulted in more rapid reduction of fluorescence signals for mismatched Let-7c than that of perfectly matched Let-7a. Behavioral discrepancy between Let-7a and Let-7c was much more apparent during surface dissociation than equilibrated hybridization. Faster releases of mismatched miRNAs from the capture DNA (compared to the perfectly matched target miRNA) can be utilized to obtain highly improved specificity for miRNA detection. Fairly consistent array signals for Let-7 miRNA detection were obtained under various dissociation conditions (Fig. 2). Note also that a sufficiently long dissociation time (1 h) was used to assure practical consistency.
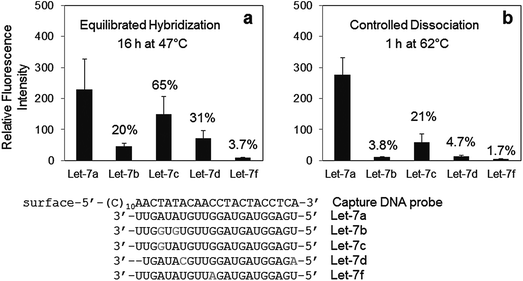
The selected dissociation condition (1 h at 62 °C) for specific Let-7a detection against Let-7c was also applied for specific discrimination of other closely related sequences of Let-7 family of miRNAs. As shown in Fig. 3, cross-hybridization levels of all other Let-7 miRNAs to the Let-7a probe were significantly reduced by a simple dissociation step. Compared with stringent equilibrium hybridization at 47 °C, our surface dissociation strategy provided up to 6-fold better specificity for Let-7a detection among four other Let-7 family members. Discrimination patterns of all Let-7 miRNAs also agreed well with a previous report.15 Single-base mismatched Let-7c showed the highest cross-hybridization with the Let-7a specific probe. On the other hand, Let-7f, with its only mismatch located in the middle of the miRNA, was better discriminated than other Let-7 family miRNAs with more mismatches such as Let-7b and Let-7d. Location of mismatches is one of the major factors affecting discrimination of short miRNAs.
General applications of controlled dissociation for specific miRNA detection
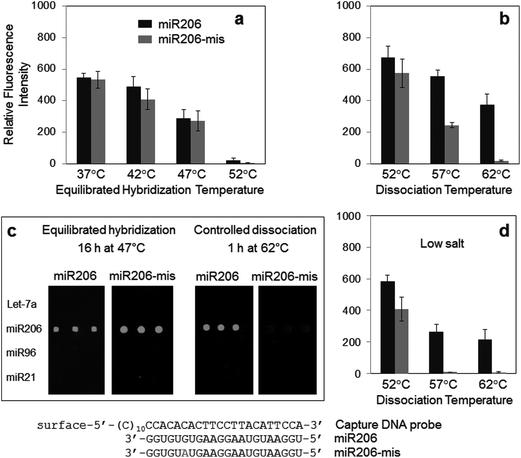
Specific miRNA detection by dissociation control was also investigated with other DNA capture probes to further demonstrate the general applicability of the method (Fig. 4). Mismatched miRNAs were designed to display similar binding properties to DNA capture probes. A single base mismatch was introduced to miR206 at the 3′ end side with a purine-to-purine substitution (miR206-mis), similar to the sequence difference between Let-7a and Let-7c (Table 1). Almost no discrimination between miR206 and miR206-mis was observed by near equilibrated hybridization at various temperatures, indicating highly similar bindings of both miRNAs to the capture probe (Fig. 4a). Surprisingly, however, the fluorescence signal of mismatched miR206-mis was dramatically lowered by stringent surface dissociation while maintaining high miR206 signals (Fig. 4b and c). Only ∼5.2% cross-hybridization was detected after 1 h dissociation at 62 °C. Considering that dissociation control can provide more diverse stringency (for example, washing temperatures and durations) to the array, somewhat improved specificity by optimizing washing conditions can be envisioned. Nonetheless, observed dissociation discrepancy between miR206 and miR206-mis, and resulting target specificity, was unexpectedly high. For highly specific surface detection of nucleic acids such as miRNAs, controlling dissociation processes is clearly more effective than controlling stringency during conventional hybridization.Dissociation stringency was also varied by the salt concentration as well as the washing duration (Fig. 4d and S1 in the ESI†). In one case, the signal of mismatched miR206 was reduced to nearly the background level (1 h dissociation at 57 °C in low salt concentration) (Fig. 4d). Lowering the salt concentration of the washing solution effectively increased the dissociation stringency. Depending on the target miRNAs, the dissociation temperature, duration, and salt concentration can be varied to select optimal washing conditions for specific miRNA detection.
Specific miRNA profiling by surface dissociation control
The practical use of complementary DNA capture probes in multiplex miRNA detection has been limited by variable melting temperatures against target miRNAs. In the present study, highly improved specificity for both Let-7a and miR206 was obtained by the same dissociation condition (62 °C for 1 h). However, the signals for other miRNAs such as miR96 and miR21 were vastly diminished under this highly stringent condition due to their relatively low melting temperatures (Fig. S2, ESI†). Washing stringency was lowered for determination of the optimal dissociation conditions for specific miR96 detection. Following 37 °C hybridization of miR96 and single-base mismatched miR96 (miR96-mis), 1 h dissociation at 57 °C afforded high miR96 signals with effectively lowered signals for miR96-mis, yielding only ∼11% cross-hybridization (Fig. S3, ESI†). Improved target specificity by controlled dissociation also yields clear signal differences between various miRNAs with diverse melting temperatures.To test our dissociation control strategy for simultaneous detection of multiple miRNAs with high specificity, we designed four LNA modified capture probes, where the melting temperatures were partially equalized (Table 1). More stable bindings of LNA probes with miRNAs required higher stringency for specific miRNA detection. Optimal dissociation conditions for these equalized LNA probes were determined based on specific Let-7a detection against Let-7c (Fig. S4, ESI†). miRNAs were first hybridized to LNA probes at 42 °C for 16 h and subsequently dissociated at 64 °C for 1 h in solution with low salt concentration. As shown in Fig. 5, all four miRNAs were effectively detected under this dissociation condition. More importantly, single base mismatched miRNAs of all four target miRNAs were discriminated with nearly background levels of cross-hybridization. These results encouragingly indicate that many other Tm equalized capture probes for miRNAs can be utilized with our dissociation strategy for better specificity of miRNA profiling experiments.
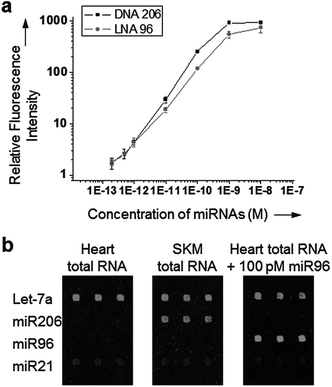
We also examined detection ranges of our dissociation controlled hybridization for target miRNAs. A wide range of miRNA concentrations (200 fM to 10 nM) was tested for two miRNAs, miR206 and miR96. DNA 206 and LNA 96 probes were used to investigate both DNA and LNA probes. A good signal linearity was observed over a concentration range of 200 fM to 1 nM for all miRNAs with a detection limit of approximately 500 fM (Fig. 6a). Detection limits and dynamic ranges were very similar to those obtained with commercially available LNA probes (Exiqon), while single-mismatch discrimination was clearly improved in our method.15 Finally, Tm equalized LNA capture probes were used to measure the expression patterns of four miRNAs in human tissue total RNAs. Human skeletal muscle (SKM) or heart total RNA extract (1 μg) was labeled with a Hi-Power Labeling kit (Exiqon) and applied to the LNA probe-spotted glass slide (Fig. 6b). Expression profiles in both RNA samples were consistent with a previous experiment.15 Based on the signal of spiked-in miR96 (100 pM), estimated concentrations of Let-7a and miR206 (20–10 pM) were also correlated well with previous reports.13,16
Experimental
Materials
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide (EDC), N-hydroxysuccinimide (NHS), succinic anhydride, and Cy3-labeled streptavidin proteins were purchased from Sigma-Aldrich. Modified DNA oligonucleotide capture probes were synthesized by Integrated DNA technologies Inc. (IDT Inc.). LNA modified probes and Hi-Power Labeling kit were purchased from Exiqon. Biotin modified miRNAs were obtained from Bioneer Inc. Amine functionalized glass slides were purchased from Nanocs. Amine-modified dextran was purchased from Invitrogen. RNase inhibitor was obtained from Roche. Total RNA extract from human heart and skeletal muscle (SKM) was purchased from Ambion.Nucleic acid capture probe design
DNA oligonucleotide capture probes are designed to recognize entire sequences of target miRNAs (Table 1). An amino linker was added to the 5′ end of all capture probes for surface immobilization, and ten cytosine nucleotides (C10) were also added between the amino linker and target recognizing sequences. Locked nucleic acid (LNA) modified capture probes were designed to interact with target miRNAs with partially equalized melting temperatures (Table 1). LNA capture probes recognize 16 bases of target miRNAs, and LNA modification was introduced in a manner that G/C bases and LNA modified bases were evenly distributed throughout the capture probes. Although calculated melting temperatures of all LNA capture probes (based on the manufacturer's program) are similar, the miR21-specific LNA probe offered relatively weaker binding to the target miRNA (miR21) than those of other LNA probes. All capture probes were dissolved in a spotting solution containing 3× SSC (450 mM NaCl and 45 mM sodium citrate) and 0.04% SDS for surface immobilization.Target microRNAs
Four synthetic human Let-7 family miRNAs (Let-7b, -c, -d, and -f) were investigated to determine their cross-hybridization to Let-7a specific probes. Other miRNAs (miR21, miR96, and miR206) and mismatched miRNAs were also investigated. Single base mismatches on miR21, miR96, and miR206 were introduced at the 3′ end sides of these miRNAs, similar to the sequence difference between Let-7a and Let-7c. Purine-to-purine or pyrimidine-to-pyrimidine substitutions were introduced for high sequence similarities and low discrimination during array hybridization. All miRNAs were synthesized with a biotin moiety for surface detection with fluorescence dye-labeled streptavidin.Microarray fabrication
Amine functionalized glass slides were incubated with 1 M succinic anhydride in DMF at 37 °C overnight to prepare carboxyl-functionalized glass slides. The resulting carboxyl glasses were activated by incubation in a mixture of 0.1 M EDC and 0.025 M NHS for 10 min. Amine modified capture probes (DNA as well as LNA modified probes) were diluted to a final concentration of 10 μM in a spotting solution (3× SSC, 0.04% SDS), and then spotted onto this NHS-coated chip. The spotted glass slides were incubated in a humid environment at room temperature for 1 h, and then washed with 0.2% SDS. After 1 h incubation, the arrayed slides were submerged in 10 mL of blocking solution I (0.1 mg mL−1 amine–dextran and 1 M ethanolamine, pH 8.5) for 1 h to block the unprinted surface.Array detection of miRNAs
Hybridization of biotinylated miRNAs with surface-immobilized capture probes was performed in a hybridization chamber (Agilent). Various miRNAs in 0.1 mL of hybridization solution (5× SSC, 0.1% SDS and 20 units RNase inhibitor) were incubated with the arrayed glass slide in a rotator hybridization incubator (Agilent) for 16 h (near equilibrated hybridization). To optimize hybridization conditions, a sufficiently high level of miRNA (100 pM) was applied. Following hybridization, hybridized glass slides were washed with a washing buffer (2× SSC with 0.1% SDS; 10 mL) in a shaking incubator at different temperatures for different periods of time, as indicated. Dissociation stringency was also increased by lowering the salt concentration to 1× SSC (150 mM NaCl and 15 mM sodium citrate) with 0.1% SDS. All washing solutions were pre-warmed at indicated temperatures for 16 h prior to use. For conventional equilibrated hybridization, hybridized glass slides were only briefly rinsed with a washing buffer (2× SSC with 0.1% SDS; 10 mL) at the temperature that was used during equilibrated hybridization.The resulting (washed) arrays were additionally blocked with a PBS solution containing 1 mg mL−1 BSA for 30 min. Biotinylated miRNAs were reacted with 1 μg mL−1 of Cy3-streptavidin protein containing 0.5 mg mL−1 BSA for 30 min. Finally, array slides were washed with PBST (PBS with 0.05% Tween 20), 0.1× PBS, and subsequently dried with nitrogen gas. Slides were analyzed with a fluorescence scanner (GenePix 4200A, Axon Instruments) at constant power and PMT gain settings (532 nm wavelength). The relative signal intensities for target miRNAs were calculated by dividing the fluorescence intensity of each miRNA spot by the same capture probe spot without target miRNA. All experiments were performed at least three independent times, and standard deviations were indicated as error bars in the graphs.
For total RNA analysis, RNAs (1 μg) were labeled with the Hi-Power Labeling kit (Exiqon), following the manufacturer's protocol. Intact synthetic miR96 (without biotin) was spiked in before labeling.
Conclusions
We demonstrated that behavioral (surface hybridization or dissociation) disparities between sequentially similar miRNAs are greatly more apparent during dissociation from capture probes than during equilibrated hybridization with these capture probes. By employing this interesting feature of miRNA hybridization and dissociation, a generally applicable hybridization strategy was introduced for miRNA surface detection with highly improved specificity. Mismatched miRNAs were much more rapidly dissociated from the capture probes than perfectly matched miRNAs, during short but highly stringent incubation. This different surface dissociation of captured miRNAs afforded surprisingly effective discrimination of single nucleotide differences in multiple miRNAs. Stringency control during conventional equilibrated hybridization was, however, not effective for differentiating these similar miRNAs. Controlling dissociation of captured miRNAs offers a simple but highly effective way to improve miRNA detection specificity. Importantly, our miRNA hybridization strategy can be easily applied to many already developed nucleic acid capture probes for improved specificity, by simply modifying the washing steps. Additionally, we are currently investigating if specificity of cellular detection of fixed miRNAs by in situ hybridization can also be improved by our hybridization strategy.Acknowledgements
This research is supported by grants from the National Research Foundation of Korea (NRF 2011-0015295), the Ministry of Science, ICT & Future Planning (2005-2001321), and a grant from the KRIBB Initiative Research Program (KRIBB, Korea).Notes and references
- D. P. Bartel, Cell, 2009, 136, 215–233 CrossRef CAS PubMed.
- J. Zhang, H. Zhao, Y. Gao and W. Zhang, Biochim. Biophys. Acta, 2012, 1826, 32–43 CAS.
- M. A. Cortez, C. Bueso-Ramos, J. Ferdin, G. Lopez-Berestein, A. K. Sood and G. A. Calin, Nat. Rev. Clin. Oncol., 2011, 8, 467–477 CrossRef CAS PubMed.
- S. Gilad, E. Meiri, Y. Yogev, S. Benjamin, D. Lebanony, N. Yerushalmi, H. Benjamin, M. Kushnir, H. Cholakh, N. Melamed, Z. Bentwich, M. Hod, Y. Goren and A. Chajut, PLoS One, 2008, 3, e3148 Search PubMed.
- M. Baker, Nat. Methods, 2010, 7, 687–692 CrossRef CAS PubMed.
- M. Castoldi, S. Schmidt, V. Benes, M. W. Hentze and M. U. Muckenthaler, Nat. Protoc., 2008, 3, 321–329 CrossRef CAS PubMed.
- S. Fang, H. J. Lee, A. W. Wark and R. M. Corn, J. Am. Chem. Soc., 2006, 128, 14044–14046 CrossRef CAS PubMed.
- J. M. Lee, H. Cho and Y. Jung, Angew. Chem., Int. Ed., 2010, 49, 8662–8665 CrossRef CAS PubMed.
- A. W. Wark, H. J. Lee and R. M. Corn, Angew. Chem., Int. Ed., 2008, 47, 644–652 CrossRef CAS PubMed.
- F. Sato, S. Tsuchiya, K. Terasawa and G. Tsujimoto, PLoS One, 2009, 4, e5540 Search PubMed.
- P. T. Nelson, W. X. Wang, B. R. Wilfred and G. Tang, Biochim. Biophys. Acta, 2008, 1779, 758–765 CrossRef CAS PubMed.
- A. Git, H. Dvinge, M. Salmon-Divon, M. Osborne, C. Kutter, J. Hadfield, P. Bertone and C. Caldas, RNA, 2010, 16, 991–1006 CrossRef CAS PubMed.
- H. Wang, R. A. Ach and B. Curry, RNA, 2007, 13, 151–159 CrossRef CAS PubMed.
- D. Duan, K. X. Zheng, Y. Shen, R. Cao, L. Jiang, Z. Lu, X. Yan and J. Li, Nucleic Acids Res., 2011, 39, e154 CrossRef CAS PubMed.
- J. M. Lee and Y. Jung, Angew. Chem., Int. Ed., 2011, 50, 12487–12490 CrossRef CAS PubMed.
- L. Neely, S. Patel, J. Garver, M. Gallo, M. Hackett, S. McLaughlin, M. Nadel, J. Harris, S. Gullans and J. Rooke, Nat. Methods, 2006, 3, 41–46 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3an01688a |
| This journal is © The Royal Society of Chemistry 2014 |