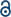 Open Access Article
Open Access ArticleBinding studies between cytosinpeptidemycin and the superfamily 1 helicase protein of tobacco mosaic virus
Xiangyang Li *,
Kai Chen,
Di Gao,
Dongmei Wang,
Maoxi Huang,
Hengmin Zhu and
Jinxin Kang
*,
Kai Chen,
Di Gao,
Dongmei Wang,
Maoxi Huang,
Hengmin Zhu and
Jinxin Kang
State Key Laboratory Breeding Base of Green Pesticide and Agricultural Bioengineering, Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Guiyang 550025, PR China. E-mail: xyli1@gzu.edu.cn; Fax: +86-851-88292090; Tel: +86-851-88292090
First published on 23rd May 2018
Abstract
Tobacco mosaic virus (TMV) helicases play important roles in viral multiplication and interactions with host organisms. They can also be targeted by antiviral agents. Cytosinpeptidemycin has a good control effect against TMV. However, the mechanism of action is unclear. In this study, we expressed and purified TMV superfamily 1 helicase (TMV-Hel) and analyzed its three-dimensional structure. Furthermore, the binding interactions of TMV-Hel and cytosinpeptidemycin were studied. Microscale thermophoresis and isothermal titration calorimetry experiments showed that cytosinpeptidemycin bound to TMV-Hel with a dissociation constant of 0.24–0.44 μM. Docking studies provided further insights into the interaction of cytosinpeptidemycin with the His375 of TMV-Hel. Mutational and Microscale thermophoresis analyses showed that cytosinpeptidemycin bound to a TMV-Hel mutant (H375A) with a dissociation constant of 14.5 μM. Thus, His375 may be the important binding site for cytosinpeptidemycin. The data are important for designing and synthesizing new effective antiphytoviral agents.
1. Introduction
Superfamily 1 (SF1) helicases are encoded in the small and large subunits of tobacco mosaic virus (TMV) and tomato mosaic virus (ToMV).1 They play important roles in viral replication. After infection, the genomic RNA is translated to produce the SF1 helicase in the cytoplasm of the host cell.2 During viral-RNA replication and control of host defenses, the SF1 helicase interacts with itself3 and with host proteins, such as TOM1, ARL8 and auxin/indole-3-acetic acid-related proteins.4–8 Then, the resistance genes N9 and Tm-1 (ref. 10 and 11) are produced and bind replication proteins to inhibit replication complex formation.12–14The ToMV SF1 helicases' (ToMV-Hel) structure, which contains two RecA domains and an N-terminal accessory domain, was determined using X-ray crystallography.15 In the ToMV-Hel N-terminal accessory domain, a long α-helix binds to TOM1 and ARL8 (ref. 4–6) for viral replication. The primary amino acid sequence of TMV helicases (TMV-Hel) have high levels of similarity to that of ToMV-Hel. The TMV-Hel and ToMV-Hel belong to Tobamovirus.16 The TMV-Hel and ToMV-Hel are also highly similar in secondary structure and function.17 Therefore, TMV-Hel is an important protein and may be a target for antiphytoviral compounds.
Cytosinpeptidemycin is an antiphytoviral antibiotic. Zhu et al. (2005) reported that cytosinpeptidemycin showed an 82.6% protection activity and 95.3% inactivate activity against TMV in tobacco. Zhu et al., using 3H-uridine and 3H-leucine tracing, found that the mechanism of cytosinpeptidemycin against TMV involved the synthesis of TMV RNA.18,19 Li et al. reported that cytosinpeptidemycin could inhibit the synthesis of the TMV methyltransferase-guanylyltransferase-like domain (at the N-terminus) and helicase-like domain (at the C-terminus) proteins using western blotting of BY-2 tobacco protoplasts. Furthermore, cytosinpeptidemycin could inhibit TMV RNA synthesis as shown in northern blots.20 However, the site of action on TMV RNA by cytosinpeptidemycin is unclear.
Because of its importance in TMV-associated disease transmission, TMV-Hel was expressed and regarded as a potential target for cytosinpeptidemycin. Here, we report binding studies using cytosinpeptidemycin and the TMV-Hel protein. Microscale thermophoresis (MST) studies revealed that the binding affinity of cytosinpeptidemycin for the TMV-Hel protein was strong, with a dissociation constant of 0.44 μM. Molecular dynamics and mutagenesis studies revealed that His375 in TMV-Hel was an important binding site for cytosinpeptidemycin. Our results demonstrated that cytosinpeptidemycin had an affinity for the TMV-Hel protein through a direct interaction with residue His375. To the best of our knowledge, this is the first report explaining the mechanism of cytosinpeptidemycin, which shows an antiviral activity against TMV.
2. Materials and methods
2.1. Drug and virus
Cytosinpeptidemycin (30%), original agent, was bought from an insecticide factory. The structural formula of compound is list in Table 1. Tobacco mosaic virus was preserved in tobacco K326 in our laboratory.2.2. RNA extraction, RT-PCR and plasmid constructs
TMV total RNA was extracted using the RNA TRIzol reagent (TaKaRa, Japan), and the total RNA was detected using a Genequant100 (GE Healthcare, USA). The cDNA synthesis methods were described in a previous report.21 The gene sequence (GenBank accession no. AJ011933.1) for TMV-Hel was used to design primers containing a Not I (underlined) restriction site in the forward primer (5′-ATAAGAATGCGGCCGCATGTCCTATTCTAAG-3′) and a XhoI (underlined) site in the reverse primer (5′-CCGCTCGAGTTAATCGACCTTATACATATC-3′). The full-length TMV-Hel gene was inserted into the vector pMCSG19 (Novagen, USA), resulting in the prokaryotic expression plasmid PMCSG19-TMV-Hel. The TMV-Hel mutant (H375A) was cloned and confirmed by 1% agarose gel electrophoresis and DNA sequencing (Shanghai Sangon Company, Shanghai, China).2.3. Protein expression and purification
A freshly transformed overnight culture of Escherichia coli strain Rosetta(DE3) (Novagen, USA) containing the plasmid pMCSG19-TMV-Hel was transferred to 1 L Luria broth supplemented with 50 μg mL−1 ampicillin at 37 °C until the OD600 reached 0.6–1.0. Then, protein expression was induced with 0.8 mM isopropyl-β-D-galactopyranoside at 16 °C overnight. The cells were removed, immediately placed in centrifuge tubes and immersed in liquid nitrogen. The cells were harvested, resuspended in lysis buffer (50 mM Tris–HCl, 150 mM NaCl and 1 mM β-mercaptoethanol, pH 7.5) and then lysed at 4 °C by sonication.22 The lysate was clarified by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 30 min at 4 °C, the soluble supernatant was loaded onto a 5 mL His-Trap high-performance column (GE Healthcare), and the protein was eluted with a linear gradient of 50–400 mM imidazole (pH 7.5). Eluate fractions collected from the column were analyzed by 12% SDS-PAGE, and the crude protein was isolated at 4 °C using a desalting column (GE Healthcare) attached to an AKTA purifier protein liquid chromatography system (GE Healthcare). The fractions containing target proteins were pooled, concentrated to a suitable concentration by ultrafiltration (10 kDa cut-off), and the MBP-tag was digested by TEV protease at 4 °C (1 mg TEV protease with 100 mg MBP-tagged fusion protein). The protein concentration was determined using a Genequant100 system (GE Healthcare), and the protein was stored at −80 °C for further analyses.
000 g for 30 min at 4 °C, the soluble supernatant was loaded onto a 5 mL His-Trap high-performance column (GE Healthcare), and the protein was eluted with a linear gradient of 50–400 mM imidazole (pH 7.5). Eluate fractions collected from the column were analyzed by 12% SDS-PAGE, and the crude protein was isolated at 4 °C using a desalting column (GE Healthcare) attached to an AKTA purifier protein liquid chromatography system (GE Healthcare). The fractions containing target proteins were pooled, concentrated to a suitable concentration by ultrafiltration (10 kDa cut-off), and the MBP-tag was digested by TEV protease at 4 °C (1 mg TEV protease with 100 mg MBP-tagged fusion protein). The protein concentration was determined using a Genequant100 system (GE Healthcare), and the protein was stored at −80 °C for further analyses.
2.4. Size-exclusion chromatography (SEC) and PAGE analysis
The purified proteins were loaded onto the Superdex 200 10/300 GL column (GE Healthcare) of an AKTA purifier protein liquid chromatography system (GE Healthcare), and the peak protein states were estimated by comparisons with protein standards. The molecular mass standards (Bio-Rad, USA) included thyroglobulin (669 kDa), ferritin (440 kDa), bovine serum albumin (67 kDa), β-lactoglobulin (35 kDa), ribonuclease A (13.7 kDa), cytochrome (13.6 kDa), aprotinin (6.51 kDa), and vitamin B12 (1.36 kDa). The proteins were monitored by absorbance at 280 nm.The fractions under the peak were collected and analyzed by 12% SDS-PAGE at room temperature. Briefly, 20 μL sample was mixed with 20 μL 2 × loading buffer [100 mM Tris–HCl (pH 6.8), 200 mM DTT, 4% (m/v) SDS, 0.2% bromophenol blue (w/v), and 20% glycerin (v/v)]. Subsequently, 4 μL samples were loaded onto 12% gels. Electrophoresis was performed using a 1 × SDS-PAGE buffer [25 mM Tris, 250 mM glycine (pH 8.3), and 0.1% SDS (m/v)] initially at 150 V at 22 °C for 20 min, and then at 120 V at 22 °C for 55 min. After electrophoresis, gels were stained with Coomassie Brilliant Blue to identify proteins, and then destained with methanol and glacial acetic acid.
2.5. Homologous model of TMV-Hel
The homology of the complete nucleotide sequences TMV-Hel and ToMV-Hel (S666 to Q1116) was analyzed using the EMBL-EBI sequence alignment program server (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The secondary structures of TMV-Hel were predicted based on their primary amino acid sequences using the ExPASy structural program server (http://www.expasy.org/structural_bioinformatics). The structure of TMV-Hel used for the study was modeled using the homologous structure of ToMV-Hel from PDB (PDB ID: 3VKW) using the ExPASy structural information for the protein (http://www.expasy.org/structural_bioinformatics). The structural representations of TMV-Hel were generated using pyMOL software (http://www.pymol.org/).232.6. Binding analysis between cytosinpeptidemycin and TMV-Hel
The binding was calculated using MST on a Monolith NR 115 (Nano Temper Technologies, Germany). Required compounds at ranging from 0.1 mM to 2 mM were incubated with 0.1 mM of purified recombinant protein for 5 min with the Monolith NT Protein Labeling Kit Red (Nano Temper Technologies) in assay buffer (10 mM PB and 150 mM sodium chloride, pH 7.5). The sample was loaded into the NanoTemper glass capillaries and microthermophoresis was carried out using 40% LED power and 40% MST. The Kd values were calculated using the mass action equation with the NanoTemper software from duplicate reads of triplicate experiments. Data are performed presented as means ± SDs of triplicate assays. Statistical differences among the data were evaluated using NanoTemper software.24,25 Isothermal titration calorimetry (ITC)-binding experiments were performed using the ITC 200 Micro Calorimeter (GE Healthcare) at 25 °C. The buffer contained 10 mM Tris–HCl and 150 mM sodium chloride (pH 7.5). The compounds (2 mM) were titrated into TMV-Hel (0.1 mM) in a 200 μL sample cell using a 40 μL microsyringe, as follows: 0.4 μL for the first injection and 2 μL for the next 19 injections at intervals of 150 s. The integrated heat data were analyzed using the one-set-of-sites model in MicroCal Origin 7.0 according to the manufacturer's instructions. Data are presented as means ± SDs of triplicate assays. Statistical differences among the data were evaluated using MicroCal Origin 7.0 software. The first data point was not used in the analysis. The binding parameters reaction enthalpy changes in cal mol−1 (ΔH), binding constant in mol−1 (K) and number of molecules per TMV-Hel (n) were floating during the fit. The binding free energy (ΔG) and reaction entropy (ΔS) were calculated using the following equations:
ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K (R = 1.9872 cal mol−1 K−1, T = 298 K), K (R = 1.9872 cal mol−1 K−1, T = 298 K),
| (1) |
| ΔG = ΔH − TΔS. | (2) |
The dissociation constant Kd was calculated as 1/K.26
2.7. Molecular docking of cytosinpeptidemycin and TMV-Hel
The initial structure of TMV-Hel was revised by adding lost residues and hydrogen atoms, and by confirming bonds and bumps. Subsequently, energy was minimized using the steepest descent calculations and conjugated gradient calculations using Discovery studio 4.5 (Accelrys, USA).27 Optimized geometries were used to construct the entire structures. The final structures were optimized by fixing the macrocycle with a conjugated gradient.The atomic Gasteiger-Huckel charges were assigned to the ligand and receptor. Most of the parameters for the docking calculation were set to the default values recommended by the software. Each docked structure was scored by the built-in scoring function and was clustered by 0.8 Å of RMSD criteria.28 For each binding model, a molecular mechanics/Poisson–Boltzmann surface area analysis was performed. Before these calculations were made, the complex structure was further refined initially with the steepest descent algorithm and then the conjugated gradient algorithm using the Amber9 package.29 During the energy minimization process, the receptor was first fixed and only the ligand remained free. Then, the ligand and residue side chains remained free. Finally, all atoms of the system were liberated and refined to a convergence of 0.01 kcal (mol−1 Å−1).
2.8. Verifying the interaction between cytosinpeptidemycin and the mutated TMV-Hel
To verify the results of the molecular docking results, a mutation, H375A, was introduced into TMV-Hel. The mutant proteins were used to measure the interactions between cytosinpeptidemycin and TMV-Hel, as previously described.24–263. Results and discussion
3.1. Molecular cloning and sequence analysis of TMV-Hel
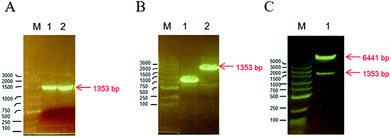
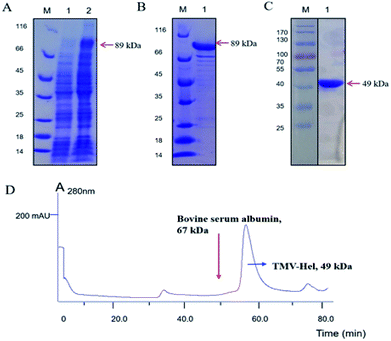
The TMV-Hel protein was cloned into pMCSG19 and highly expressed in E. coli. The TMV-Hel protein from the TMV replication protein's S1 to Q451 segment (corresponding to S666 to Q1116) was successfully amplified using specific primers and cloned into the pMCSG19 vector (Fig. 1). A sequence analysis showed that TMV-Hel (isolated from Guizhou Province) consisted of 1353 nt and shared a 99% nucleotide identity with the published sequence of a TMV replication protein isolates in National Center of Biotechnology Information protein database (GenBank accession no. AJ011933.1).3.2. Expression and purification of TMV-Hel
To obtain sufficient quantities of TMV-Hel for biochemical analyses, the expression vector pMCSG19 containing the TMV-Hel gene was transformed into E. coli strain Rosetta (DE3). A large number of TMV-Hel genes were overexpressed when the final isopropyl-β-D-galactopyranoside concentration was increased to 0.8 mM and the solution was left overnight at 16 °C (Fig. 2A and B). The His-MBP-tagged fusion protein was purified using a Ni-NTA column (GE Healthcare). More than 90% of the TMV-Hel-MBP protein was eluted from the beads with 400 mM imidazole in Tris–HCl buffer (pH 7.5), and an 89 kDa protein was revealed by 12% SDS-PAGE analysis (Fig. 2C). The final yield from 500 mL of bacterial culture was ∼0.5 mg of purified TMV-Hel-MBP protein after using a desalting column. Then, MBP tags were digested by TEV protease to produce TMV-Hel protein.3.3. The aggregating state of TMV-Hel
To determine the aggregating state of TMV-Hel in vitro, fresh TMV-Hel was isolated and analyzed by size-exclusion chromatograph. The TMV-Hel eluted in the corresponding position of 67 kDa in 10 mM Tris–HCl and 150 mM sodium chloride (pH 7.5) (Fig. 2D). Thus, the monomer form of TMV-Hel was ∼49 kDa.3.4. Interactions between cytosinpeptidemycin and TMV-Hel
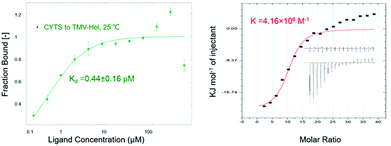
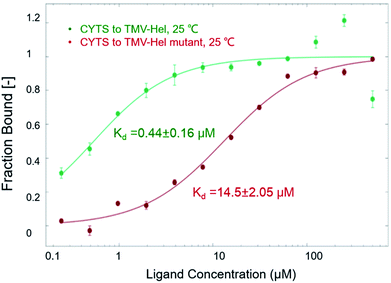
The binding interactions between cytosinpeptidemycin and TMV-Hel were analyzed by MST. Cytosinpeptidemycin bound to TMV-Hel protein with a Kd of 0.44 ± 0.16 μM (Fig. 3A). Analysis by ITC revealed that one TMV-Hel interacted with three cytosinpeptidemycin molecules, and cytosinpeptidemycin bound to TMV-Hel with a K of 4.16 ± 2.23 × 106 M−1 (Fig. 3B). The titration data also indicated an apparent negative enthalpy value (ΔG = −1.74 × 107 kJ mol−1) during the binding of cytosinpeptidemycin to TMV-Hel. Cytosinpeptidemycin had a micromole affinity with TMV-Hel.3.5. Homology modelling of TMV-Hel
A BLAST algorithm-based analysis using the EMBL-EBI sequence alignment program server, found an 89.6% homology between TMV-Hel and ToMV-Hel. Based on their primary amino acid sequences, the TMV-Hel protein's secondary structures were predicted, as shown in Fig. 4. The homology structure of TMV-Hel was modelled using the X-ray crystal structure of ToMV-Hel (PDB ID: 3VKW). The structural representation of TMV helicase is shown in Fig. 5.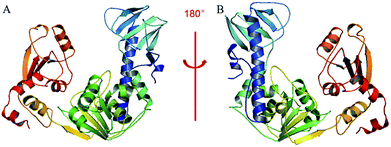 | ||
| Fig. 5 Based on the X-ray crystal structure of ToMV-Hel (PDB code: 3VKW), the TMV-Hel was modeled. (A) TMV-Hel; (B) was rotate 180 degrees of (A). The figures for structural representation of TMV-Hel were generated on pyMOL software. | ||
3.6. Molecular docking of cytosinpeptidemycin and TMV-Hel
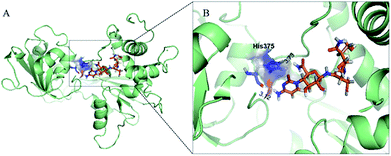
To identify the cytosinpeptidemycin recognition sites in TMV-Hel, we performed molecular docking using the Gold method with 200 cycles. After optimization, we found that His375 in the TMV-Hel shared two hydrogen bonds with cytosinpeptidemycin, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N = 3.12 Å and H–N⋯H–N = 3.73 Å (Fig. 6). The BLAST algorithm-based analysis revealed that TMV-Hel shared a high homology level with known ToMV-Hel protein in GenBank, with His375 being present in a highly conserved region (Fig. 4). His375 (named His1040 in ToMV-Hel) is an important residue for TMV-Hel nucleic acid binding.30 These analyses indicated that His375 is the reason for the decreasing activity level of cytosinpeptidemycin.
O⋯H–N = 3.12 Å and H–N⋯H–N = 3.73 Å (Fig. 6). The BLAST algorithm-based analysis revealed that TMV-Hel shared a high homology level with known ToMV-Hel protein in GenBank, with His375 being present in a highly conserved region (Fig. 4). His375 (named His1040 in ToMV-Hel) is an important residue for TMV-Hel nucleic acid binding.30 These analyses indicated that His375 is the reason for the decreasing activity level of cytosinpeptidemycin.
3.7. Mutational analysis of the protein–ligand interactions between cytosinpeptidemycin and TMV-Hel
To analyze the roles of the critical residue in the binding of TMV-Hel by cytosinpeptidemycin that was identified in the simulations, we cloned and expressed a TMV-Hel mutant, H375A. The interactions between the mutant and cytosinpeptidemycin were studied using MST, the affinity level between the TMV-Hel mutant and cytosinpeptidemycin decreased (14.5 ± 2.05 μM) compared with the wild type (0.44 ± 0.16 μM) (Fig. 7). Thus, we deduced that His375 is the binding site of TMV-Hel.3.8. The putative reaction mechanism of cytosinpeptidemycin on TMV-Hel
Recent reports showed that the ToMV-Hel's conserved helicase motifs had NTP and nucleic acid-binding sites.31–35 The high homology of TMV-Hel with ToMV-Hel indicates that they may have similar functions. Furthermore, cytosinpeptidemycin could inhibit the synthesis of TMV-Hel as shown by western blotting in BY-2 tobacco protoplasts,20 and TMV-Hel's His375 is associated with RNA binding.30 The residue His375 was contained in a highly conserved region in both TMV-Hel and ToMV-Hel; therefore, the biological function of His375 in TMV-Hel could also be associated with RNA binding. Our experiments demonstrated that His375 was also the binding site of cytosinpeptidemycin. Based on these results, we hypothesize that cytosinpeptidemycin inhibited the virulence of TMV through its interaction with His375.4. Conclusions
In summary, our study aimed to explain the mechanism of action of cytosinpeptidemycin against TMV-Hel. First, we expressed TMV-Hel and constructed a structural model as a potential target for antiphytoviral compounds. Second, we found that cytosinpeptidemycin had a micromole affinity for TMV-Hel. Last, we confirmed that cytosinpeptidemycin bound His375 in the TMV-Hel protein, which was important for the binding of TMV RNA. Thus, cytosinpeptidemycin inhibited the virulence of TMV in vitro by binding TMV-Hel via chemical bonds. This competition for the residues contributed to viral replication. Our results indicate, for the first time, that cytosinpeptidemycin inhibits TMV infection by binding His375 in the TMV-Hel protein.To the best of our knowledge, this is the first reported explanation of the mechanism for cytosinpeptidemycin that shows antiviral activity against TMV-Hel, and this study also establishes a useful model system that allows the rapid screening of antiviral agents.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 21502032 and No. 31460460).References
- G. Kadare and A. L. Haenni, Virus-encoded RNA helicases, J. Virol., 1997, 71, 2583–2590 Search PubMed.
- S. P. Goregaoker, D. J. Lewandowski and J. N. Culver, Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus, Virology, 2001, 282, 320–328 CrossRef PubMed.
- K. Ishibashi, M. Nishikiori and M. Ishikawa, Interactions between tobamovirus replication proteins and cellular factors: their impacts on virus multiplication, Mol. Plant-Microbe Interact., 2010, 23, 1413–1419 CrossRef PubMed.
- M. Nishikiori, M. Mori, K. Dohi, H. Okamura, E. Katoh, S. Naito, T. Meshi and M. Ishikawa, A host small GTP-binding protein ARL8 plays crucial roles in Tobamovirus RNA replication, PLoS Pathog., 2011, 7, e1002409 Search PubMed.
- T. Yamanaka, T. Imai, R. Satoh, A. Kawashima, M. Takahashi, K. Tomita, K. Kubota, T. Meshi, S. Naito and M. Ishikawa, Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes, J. Virol., 2002, 76, 2494–2497 CrossRef.
- T. Yamanaka, T. Ohta, M. Takahashi, T. Meshi, R. Schmidt, C. Dean, S. Naito and M. Ishikawa, TOM1, an arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 10107–10112 CrossRef PubMed.
- M. S. Padmanabhan, S. P. Goregaoker, S. Golem, H. Shiferaw and J. N. Culver, Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development, J. Virol., 2005, 79, 2549–2558 CrossRef PubMed.
- M. S. Padmanabhan, S. R. Kramer, X. Wang and J. N. Culver, Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection, J. Virol., 2008, 82, 2477–2485 CrossRef PubMed.
- H. Ueda, Y. Yamaguchi and H. Sano, Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants, Plant Mol. Biol., 2006, 61, 31–45 CrossRef PubMed.
- K. Ishibashi, K. Masuda, S. Naito, T. Meshi and M. Ishikawa, An inhibitor of viral RNA replication is encoded by a plant resistance gene, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13833–13838 CrossRef PubMed.
- M. Kato, K. Ishibashi, C. Kobayashi, M. Ishikawa and E. Katoh, Expression, purification, and functional characterization of an n-terminal fragment of the tomato mosaic virus resistance protein tm-1, Protein Expression Purif., 2013, 89, 1–6 CrossRef PubMed.
- K. Ishibashi, Y. Kezuka, C. Kobayashi, M. Kato, T. Inoue, T. Nonaka, M. Ishikawa, H. Matsumura and E. Katoh, Structural basis for the recognition-evasion arms race between tomato mosaic virus and the resistance gene tm-1, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3486–3495 CrossRef PubMed.
- K. Ishibashi and M. Ishikawa, The Resistance Protein Tm-1 Inhibits Formation of a Tomato Mosaic Virus Replication Protein-Host Membrane Protein Complex, J. Virol., 2013, 87, 7933–7939 CrossRef PubMed.
- K. Ishibashi and M. Ishikawa, Mechanisms of tomato mosaic virus RNA replication and its inhibition by the host resistance factor Tm-1, Curr. Opin. Virol., 2014, 9, 8–13 CrossRef PubMed.
- M. Nishikiori, S. Sugiyama, H. Y. Xiang, M. Niiyama, K. Ishibashi, T. Inoue, M. Ishikawa, H. Matsumura and E. Katoh, Crystal structure of the superfamily 1 helicase from tomato mosaic virus, J. Virol., 2012, 86, 7565–7576 CrossRef PubMed.
- H. Xiang, K. Ishibashi, M. Nishikiori, M. C. Jaudal, M. Ishikawa and E. Katoh, Expression, purification, and functional characterization of a stable helicase domain from a tomato mosaic virus replication protein, Protein Expression Purif., 2012, 81, 89–95 CrossRef PubMed.
- Q. Zhou Study on molecular design, synthesis and bioactivity of insecticides based on the RNA helicase of tobacco Mosaic virus, Dissertation, Guizhou University, 2014.
- C. Y. Zhu, Y. H. Wu, X. X. Zhao and Y. H. Wang, Action mechanism of Cytosinpeptidemycin against tobacco mosaic virus, Acta Phys. Sin., 2006, 36, 314–316 Search PubMed.
- C. Y. Zhu, The Action mechanism of Cytosinpeptidemycin on tobacco mosaic virus, Dissertation, Shenyang Agricultural University, 2005.
- Y. L. Li, The Action Mechanism of Cytosinpeptidemycin on the Replication Protein Gene Expression of Tobacco Mosaic Virus, Dissertation, Shenyang Agricultural University, 2017.
- X. Y. Li, B. A Song, X. Chen, Z. C. Wang, M. J. Zeng, D. D. Yu, D. Y. Hu, Z. Chen, L. H. Jin, S. Yang, C. G. Yang and B. E. Chen, Crystal structure of a four-layer aggregate of engineered TMV CP implies the importance of terminal residues for oligomer assembly, PLoS One, 2013, 8, e77717 Search PubMed.
- X. Y. Li, G. F. Hao, Q. M. Wang, Z. Chen, Y. Ding, Y. Lu, D. Y. Hu and B. A. Song, Ningnanmycin inhibits tobacco mosaic virus virulence by binding directly to its coat protein discs, Oncotarget, 2017, 8, 82446–82458 Search PubMed.
- W. L. DeLano, The pymol molecular graphics system 2010 Search PubMed.
- X. Y. Li, J. Liu, X. Yang, Y. Ding, J. Wu, D. Y. Hu and B. A. Song, Studies of binding interactions between Dufulin and southern rice black-streaked dwarf virus P9-1, Bioorg. Med. Chem., 2015, 23, 3629–3637 CrossRef PubMed.
- C. J. Wienken, P. Baaske, U. Rothbauer, D. Braun and S. Duhr, Protein-binding assays in biological liquids using microscale thermophoresis, Nat. Commun., 2010, 1, 100 CrossRef PubMed.
- A. Das and G. S. Kumar, Binding studies of aristololactam-β-D-glucoside and daunomycin to human serum albumin, RSC Adv., 2014, 4, 33082–33090 RSC.
- J. N. Ding, Y. J. Zhang, H. Zhong, C. C. Ao, J. Li and J. G. Han, An all-atom molecular dynamics study of the anti-interferon signaling of ebola virus: interaction mechanisms of ebov vp24 binding to karyopherin alpha5, Mol. BioSyst., 2017, 13, 1031–1045 RSC.
- L. A. Bratholm and J. H. Jensen, Protein structure refinement using a quantum mechanics-based chemical shielding predictor, Chem. Sci., 2016, 8, 2061–2072 RSC.
- F. Ntie-Kang, S. Kannan, K. Wichapong, L. C. Owono Owono, W. Sippl and E. Megnassan, Binding of pyrazole-based inhibitors to mycobacterium tuberculosis pantothenate synthetase: docking and mm-gb(pb)sa analysis, Mol. BioSyst., 2014, 10, 223–239 RSC.
- X. Wang, Z. Kelman and J. N. Culver, Helicase ATPase activity of the tobacco mosaic virus 126-kDa protein modulates replicase complex assembly, Virology, 2010, 402, 292–302 CrossRef PubMed.
- Z. Cheng, D. Muhlrad, M. K. Lim, R. Parker and H. Song, Structural and functional insights into the human Upf1 helicase core, EMBO J., 2007, 26, 253–264 CrossRef PubMed.
- S. S. Velankar, P. Soultanas, M. S. Dillingham, H. S. Subramanya and D. B. Wigley, Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism, Cell, 1999, 97, 75–84 CrossRef PubMed.
- H. Xiang, S. Sugiyama, H. Adachi, K. Takano, S. Murakami, T. Inoue, Y. Mor, M. Ishikawa and E. Katoh, Crystallization and preliminary X-ray crystallographic analysis of a helicase-like domain from a tomato mosaic virus replication protein, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2011, 67, 1649–1652 CrossRef PubMed.
- C. H. Gross and S. Shuman, Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase II, a DExH box RNA helicase, J. Virol., 1995, 69, 4727–4736 Search PubMed.
- A. Pause and N. Sonenberg, Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A, EMBO J., 1992, 11, 2643–2654 Search PubMed.
| This journal is © The Royal Society of Chemistry 2018 |